User:Ophelie Lefort/Sandbox
From Proteopedia
(Difference between revisions)
| Line 12: | Line 12: | ||
[[Image:Linear structure of PRSS57 (4Q7X).png]] | [[Image:Linear structure of PRSS57 (4Q7X).png]] | ||
| + | |||
The protein has a 283 amino-acids long sequence with two mainly domains. | The protein has a 283 amino-acids long sequence with two mainly domains. | ||
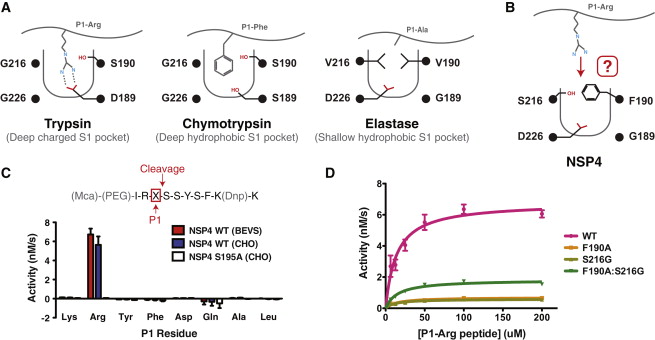
It forms a kind of elongate sphere of approximately 70Å large and 150Å long (dimensions: 70,37Å X 70,37Å X 105,02Å). The protein is composed of signal peptide (1-31) which leads to the location in azurophil granules [http://www.jimmunol.org/content/191/5/2700] and allows its excretion. In addition there is a protease domain (32-283) which is a trypsin-like domain with a trypsin-like <scene name='75/751133/Active_site/1'>active site</scene> , according to the specificity for P1-Arg residues, but this domain can be an elastase-like active site according to the primary sequence (because of the presence of a swallow S1 pocket) specific to small aliphatic residues.<ref>S.. Jack Lin, Ken C. Dong, Charles Eigenbrot, Menno van Lookeren Campagne, Daniel Kirchhofer Structures of Neutrophil Serine Protease 4 Reveal an Unusual Mechanism of Substrate Recognition by a Trypsin-Fold Protease DOI: http://dx.doi.org/10.1016/j.str.2014.07.008</ref> | It forms a kind of elongate sphere of approximately 70Å large and 150Å long (dimensions: 70,37Å X 70,37Å X 105,02Å). The protein is composed of signal peptide (1-31) which leads to the location in azurophil granules [http://www.jimmunol.org/content/191/5/2700] and allows its excretion. In addition there is a protease domain (32-283) which is a trypsin-like domain with a trypsin-like <scene name='75/751133/Active_site/1'>active site</scene> , according to the specificity for P1-Arg residues, but this domain can be an elastase-like active site according to the primary sequence (because of the presence of a swallow S1 pocket) specific to small aliphatic residues.<ref>S.. Jack Lin, Ken C. Dong, Charles Eigenbrot, Menno van Lookeren Campagne, Daniel Kirchhofer Structures of Neutrophil Serine Protease 4 Reveal an Unusual Mechanism of Substrate Recognition by a Trypsin-Fold Protease DOI: http://dx.doi.org/10.1016/j.str.2014.07.008</ref> | ||
Revision as of 09:19, 27 January 2017
Neutrophil Serine 4 or Serine Protease 57 (4Q7X)
| |||||||||||
References
[3] NSP4 is stored in azurophil granules and released by activated neutrophils as active endoprotease with restricted specificity, NCBI
[4] NSP4, an elastase-related protease in human neutrophils with arginine specificity, NCBI
[5] Tailor-made inflammation: how neutrophil serine proteases modulate the inflammatory response, NCBI
[6] Role of neutrophils in innate immunity: a systems biology-level approach, NCBI
[7] Neutrophils: Their Role in Innate and Adaptive Immunity, NCBI
[8] Neutrophil extracellular traps kill bacteria, NCBI
[9] Tailor-made inflammation: how neutrophil serine proteases modulate the inflammatory response, NCBI
- ↑ S.. Jack Lin, Ken C. Dong, Charles Eigenbrot, Menno van Lookeren Campagne, Daniel Kirchhofer Structures of Neutrophil Serine Protease 4 Reveal an Unusual Mechanism of Substrate Recognition by a Trypsin-Fold Protease DOI: http://dx.doi.org/10.1016/j.str.2014.07.008
- ↑ Natascha C. Perera, Karl-Heinz Wiesmüller, Maria Torp Larsen, Beate Schacher, Peter Eickholz, Niels Borregaard and Dieter E. Jenne NSP4 Is Stored in Azurophil Granules and Released by Activated Neutrophils as Active Endoprotease with Restricted Specificity DOI: https://doi.org/10.4049/jimmunol.1301293
- ↑ Lin, S. Jack, Ken C. Dong, Charles Eigenbrot, Menno van Lookeren Campagne, and Daniel Kirchhofer. “Structures of Neutrophil Serine Protease 4 Reveal an Unusual Mechanism of Substrate Recognition by a Trypsin-Fold Protease.” Structure 22, no. 9 (September 2, 2014): 1333–40.[1]
- ↑ Perera, Natascha C., Karl-Heinz Wiesmüller, Maria Torp Larsen, Beate Schacher, Peter Eickholz, Niels Borregaard, and Dieter E. Jenne. “NSP4 Is Stored in Azurophil Granules and Released by Activated Neutrophils as Active Endoprotease with Restricted Specificity.” The Journal of Immunology 191, no. 5 (September 1, 2013) [2]