Sandbox Reserved 1269
From Proteopedia
(→Structure, Interactions, and Location) |
|||
| Line 1: | Line 1: | ||
<Structure load='1ysa' size='350' frame='true' align='right' caption='1ysa' scene='Insert optional scene name here' /> | <Structure load='1ysa' size='350' frame='true' align='right' caption='1ysa' scene='Insert optional scene name here' /> | ||
| - | + | ==1YSA== | |
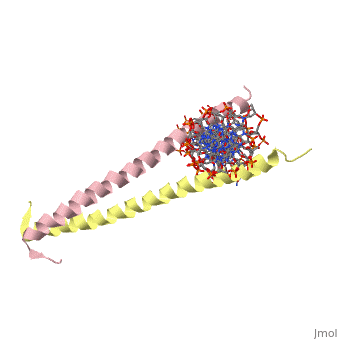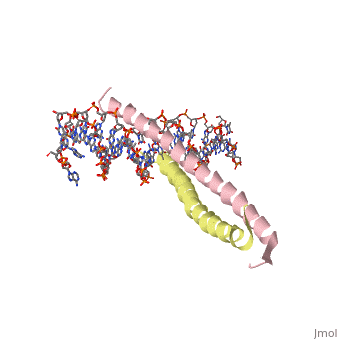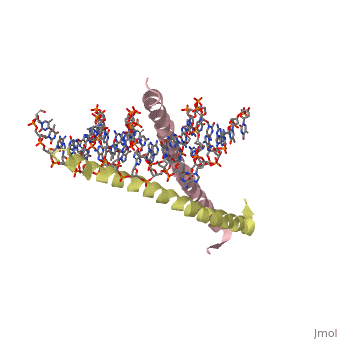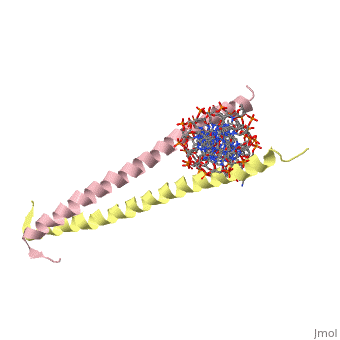
| + | 1YSA is the PDB of GCN4 leucine zipper. The zipper is coil of alpha helices which interact through ion pairs and hydrogen bonding. The leucines make side-to-side interactions in every other layer of the dimer structure that is formed with the DNA (5). | ||
==Function of 1YSA== | ==Function of 1YSA== | ||
1YSA is a transcription factor that activates over 30 genes necessary for amino acid or <scene name='75/751162/Purine/1'>purine</scene> biosynthesis. 1YSA binds to the DNA sequence: 5'-TGA[CG]TCA-3' (1). | 1YSA is a transcription factor that activates over 30 genes necessary for amino acid or <scene name='75/751162/Purine/1'>purine</scene> biosynthesis. 1YSA binds to the DNA sequence: 5'-TGA[CG]TCA-3' (1). | ||
==Structure, Interactions, and Location== | ==Structure, Interactions, and Location== | ||
| - | 1YSA is a <scene name='75/751162/Four-chain_structure/1'> | + | 1YSA is a four chain, <scene name='75/751162/Four-chain_structure/1'>two-stranded</scene> transcription factor. There are both polar and <scene name='75/751162/Nonpolar/1'>nonpolar</scene> regions within 1YSA. It interacts with DNA and binds DNA as a dimer. 1YSA interacts in several locations within the DNA. Additionally, 1YSA interacts with the protein Gcn4. Within DNA, 1YSA binds to DNA (5'-3 D(*tp*tp*cp*cp*tp*ap*tp*gp*ap*cp*tp*cp*ap*tp*cp*cp*a4 P*gp*tp*t)-3') and DNA (5'-9 D(*ap*ap*ap*cp*tp*gp*gp*ap*tp*gp*ap*gp*tp*cp*ap*tp*a10 P*gp*gp*a)-3') (2). The location of 1YSA within a cell is in nucleus P03069 (3). |
==Origin== | ==Origin== | ||
| Line 19: | Line 20: | ||
(4) www.uniprot.org/uniprot/P03069 | (4) www.uniprot.org/uniprot/P03069 | ||
| + | |||
| + | (5) www.ncbi.nlm.nih.gov/pubmed/1948029 | ||
---- | ---- | ||
Revision as of 20:58, 8 February 2017
|
Contents |
1YSA
1YSA is the PDB of GCN4 leucine zipper. The zipper is coil of alpha helices which interact through ion pairs and hydrogen bonding. The leucines make side-to-side interactions in every other layer of the dimer structure that is formed with the DNA (5).
Function of 1YSA
1YSA is a transcription factor that activates over 30 genes necessary for amino acid or biosynthesis. 1YSA binds to the DNA sequence: 5'-TGA[CG]TCA-3' (1).
Structure, Interactions, and Location
1YSA is a four chain, transcription factor. There are both polar and regions within 1YSA. It interacts with DNA and binds DNA as a dimer. 1YSA interacts in several locations within the DNA. Additionally, 1YSA interacts with the protein Gcn4. Within DNA, 1YSA binds to DNA (5'-3 D(*tp*tp*cp*cp*tp*ap*tp*gp*ap*cp*tp*cp*ap*tp*cp*cp*a4 P*gp*tp*t)-3') and DNA (5'-9 D(*ap*ap*ap*cp*tp*gp*gp*ap*tp*gp*ap*gp*tp*cp*ap*tp*a10 P*gp*gp*a)-3') (2). The location of 1YSA within a cell is in nucleus P03069 (3).
Origin
1YSA originates from the organism Saccharomyces cerevisiae, or commonly known as baker's yeast (4).
References
(1) www.proteopedia.org/wiki/index.php/1ysa
(2) www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=1ysa
(3) www.pdbj.org/mine/summary/1ysa
(4) www.uniprot.org/uniprot/P03069
(5) www.ncbi.nlm.nih.gov/pubmed/1948029