We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1051
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
===Zinc Binding === | ===Zinc Binding === | ||
Zinc acts as an inhibitor to Czr A, thus preventing transcriptional repression of Czr B and allowing Zn<sup>2+</sup> transport out of the cell. This allows for zinc transport to essentially be self regulated. That is, when zinc concentration in the cell is relatively high, zinc ions bind to Czr A, causing a conformational change which releases the bound DNA. DNA without Czr A bound is free to be transcribed and Czr B is again expressed, allowing for Zn<sup>2+</sup> transport out of the cell. | Zinc acts as an inhibitor to Czr A, thus preventing transcriptional repression of Czr B and allowing Zn<sup>2+</sup> transport out of the cell. This allows for zinc transport to essentially be self regulated. That is, when zinc concentration in the cell is relatively high, zinc ions bind to Czr A, causing a conformational change which releases the bound DNA. DNA without Czr A bound is free to be transcribed and Czr B is again expressed, allowing for Zn<sup>2+</sup> transport out of the cell. | ||
| + | |||
== Structural Overview == | == Structural Overview == | ||
| - | CzrA functions as a dimer. The <scene name='69/694218/Monomeric_unit/1'>monomeric units</scene> dimerize at the czr operon, repressing gene transcription. Each monomeric unit contains <scene name='69/694218/Helices/1'>five alpha helices</scene> seen in purple and <scene name='69/694218/B_sheets/1'>two beta sheets</scene> displayed in yellow. While the function of the beta sheets are not yet known, key helices regulate the binding of DNA and Zn<sup> +2 </sup>. The <scene name='69/694218/Alpha_4_helix/1'>alpha 4 helix</scene> is the location of DNA binding and the <scene name='69/694218/Alpha_5_helix/1'>alpha 5 helix</scene> contains the Zn<sup> +2 </sup> binding site. As Zn<sup> +2 </sup> binds, the alpha 4 helices are <scene name='69/694218/Alpha_4_helices_pushed/1'>pushed out of alignment</scene>, repressing their DNA binding ability. | + | CzrA functions as a dimer[https://en.wikipedia.org/wiki/Dimer_(chemistry)]. The <scene name='69/694218/Monomeric_unit/1'>monomeric units</scene> dimerize at the czr operon, repressing gene transcription. Each monomeric unit contains <scene name='69/694218/Helices/1'>five alpha helices</scene> seen in purple and <scene name='69/694218/B_sheets/1'>two beta sheets</scene> displayed in yellow. While the function of the beta sheets[https://en.wikipedia.org/wiki/Beta_sheet] are not yet known, key helices[https://en.wikipedia.org/wiki/Alpha_helix] regulate the binding of DNA and Zn<sup> +2 </sup>. The <scene name='69/694218/Alpha_4_helix/1'>alpha 4 helix</scene> is the location of DNA binding and the <scene name='69/694218/Alpha_5_helix/1'>alpha 5 helix</scene> contains the Zn<sup> +2 </sup> binding site. As Zn<sup> +2 </sup> binds, the alpha 4 helices are <scene name='69/694218/Alpha_4_helices_pushed/1'>pushed out of alignment</scene>, repressing their DNA binding ability. |
== Binding of DNA == | == Binding of DNA == | ||
| Line 23: | Line 24: | ||
== Zinc Binding == | == Zinc Binding == | ||
| - | Most zinc-dependent proteins are transcriptional regulators<ref>DOI: 10.1128/MMBR.00015-06</ref>. CzrA fits into this category as an allosteric inhibitor of the czr operon. Two Zn<sup> +2 </sup> ions may bind to the dimer<ref>DOI:10.1073/pnas.0636943100</ref>, at the location of the <scene name='69/694218/Alpha_5_helices/2'> alpha 5 </scene> helix from each monomer. As zinc binds, the alpha 5 helices <scene name='69/694218/2kjc_zinc_bound/1'>swing down</scene> to inhibit the DNA binding residues. Furthermore, CzrA must be in its dimer form for zinc to bind. The <scene name='69/694218/Spacefill_with_zinc_pockets/1'>zinc binding pocket</scene> is formed by two residues from each monomer, so Zn<sup>+2</sup> cannot bind to the monomer. The <scene name='69/694218/Zinc_residues/1'>zinc binding site</scene> is formed by Asp84 and His86 from one monomer, and His97 and His100 from the other monomer. Histidines are a repetitive and commonly found residue in zinc-binding proteins <ref>Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun 4;4(6):1609-1614.</ref>. | + | Most zinc-dependent proteins are transcriptional regulators<ref>DOI: 10.1128/MMBR.00015-06</ref>. CzrA fits into this category as an allosteric inhibitor[https://en.wikipedia.org/wiki/Allosteric_regulation] of the czr operon. Two Zn<sup> +2 </sup>[https://en.wikipedia.org/wiki/Zinc] ions may bind to the dimer<ref>DOI:10.1073/pnas.0636943100</ref>, at the location of the <scene name='69/694218/Alpha_5_helices/2'> alpha 5 </scene> helix from each monomer. As zinc binds, the alpha 5 helices <scene name='69/694218/2kjc_zinc_bound/1'>swing down</scene> to inhibit the DNA binding residues. Furthermore, CzrA must be in its dimer form for zinc to bind. The <scene name='69/694218/Spacefill_with_zinc_pockets/1'>zinc binding pocket</scene> is formed by two residues from each monomer, so Zn<sup>+2</sup> cannot bind to the monomer. The <scene name='69/694218/Zinc_residues/1'>zinc binding site</scene> is formed by Asp84 and His86 from one monomer, and His97 and His100 from the other monomer. Histidines are a repetitive and commonly found residue in zinc-binding proteins <ref>Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun 4;4(6):1609-1614.</ref>. |
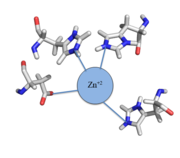
| - | Zinc<sup>+2</sup> binding is driven by a large entropic gain <ref>DOI:10.1021/ja906131b</ref>. Water molecules around the metal ion and CzrA protein are displaced, and gain greater freedom. This gain in entropy allows Zn<sup>+2</sup> to bind to CzrA with reasonable affinity and speed in vivo. The zinc<sup>+2</sup> ion forms a tetrahedral complex with the four residues (Figure 1). This allows other metal ions to act as allosteric inhibitors to CzrA. Any metal that may form a tetrahedral complex will have some affinity for CzrA, assuming it is not too large to fit into the pocket. However, the metal binding pocket of CzrA has been optimized to bind Zn<sup>+2</sup> with the highest affinity. As CzrA is a transcriptional repressor, binding of Zn<sup>+2</sup> to the dimer will activate the czr operon. Zn<sup>+2</sup> is preferred as CzrB opens a Zn<sup>+2</sup> channel, allowing the excess zinc ions to export the cell. | + | Zinc<sup>+2</sup> binding is driven by a large entropic[https://en.wikipedia.org/wiki/Entropy] gain <ref>DOI:10.1021/ja906131b</ref>. Water molecules around the metal ion and CzrA protein are displaced, and gain greater freedom. This gain in entropy allows Zn<sup>+2</sup> to bind to CzrA with reasonable affinity and speed in vivo. The zinc<sup>+2</sup> ion forms a tetrahedral complex with the four residues (Figure 1). This allows other metal ions to act as allosteric inhibitors to CzrA. Any metal that may form a tetrahedral complex will have some affinity for CzrA, assuming it is not too large to fit into the pocket. However, the metal binding pocket of CzrA has been optimized to bind Zn<sup>+2</sup> with the highest affinity. As CzrA is a transcriptional repressor, binding of Zn<sup>+2</sup> to the dimer will activate the czr operon. Zn<sup>+2</sup> is preferred as CzrB opens a Zn<sup>+2</sup> channel, allowing the excess zinc ions to export the cell. |
[[Image:Zinc tetrahedral complex.PNG|thumb|center| Figure 1:Zn<sup>+2</sup> tetrahedral binding complex]] | [[Image:Zinc tetrahedral complex.PNG|thumb|center| Figure 1:Zn<sup>+2</sup> tetrahedral binding complex]] | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 13:45, 28 March 2017
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
CzrA
| |||||||||||