Sandbox Reserved 1069
From Proteopedia
| Line 8: | Line 8: | ||
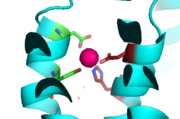
YiiP is a homodimer, with each monomer consisting of 238 residues [https://en.wikipedia.org/wiki/Protein_dimer (protein dimer)] with a transmembrane (TMD) and C-terminal (CTD) domain that are connected via a charge interlocking mechanism located on a flexible loop. There are 3 Zn<sup>2+</sup> binding sites per unit of homodimer. Site A is located in the TMD, site C is located in the CTD, and site B is located at the junction of the domains join. Both TMD are composed of 6 helices, 4 of which (TM1,TM2,TM4,TM5) form a pore in which Zn<sup>2+</sup> and H<sup>+</sup> can reach binding Site A. Zn<sup>2+</sup> binding at site C helps hold the CTD together and is thought to stabilize conformational changes in YiiP. | YiiP is a homodimer, with each monomer consisting of 238 residues [https://en.wikipedia.org/wiki/Protein_dimer (protein dimer)] with a transmembrane (TMD) and C-terminal (CTD) domain that are connected via a charge interlocking mechanism located on a flexible loop. There are 3 Zn<sup>2+</sup> binding sites per unit of homodimer. Site A is located in the TMD, site C is located in the CTD, and site B is located at the junction of the domains join. Both TMD are composed of 6 helices, 4 of which (TM1,TM2,TM4,TM5) form a pore in which Zn<sup>2+</sup> and H<sup>+</sup> can reach binding Site A. Zn<sup>2+</sup> binding at site C helps hold the CTD together and is thought to stabilize conformational changes in YiiP. | ||
| - | Each [https://en.wikipedia.org/wiki/Monomer monomer] structure is classified into two different domains, the Trans-Membrane Domain (TMD) and C-Terminus Domain (CTD). The TMD consists of a total of six helices in each | + | Each [https://en.wikipedia.org/wiki/Monomer monomer] structure is classified into two different domains, the Trans-Membrane Domain (TMD) and C-Terminus Domain (CTD). The TMD, where Zn binding site A resides, consists of a total of six helices in each <scene name='75/756372/Sixhelices/1'>monomer</scene>. Four of these helices are bundled together while the remaining two are oriented antiparallel to the <scene name='75/756372/2antiparallel/1'>bundle</scene>. Movement of these helices play a role in the function of YiiP. A large portion of the protein containing binding site C, approximately 30 Å in length<sup>[https://www.bnl.gov/isd/documents/71335.pdf]</sup>, protrudes into the cytoplasm functioning as a zinc sensor within the cell. |
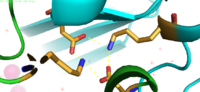
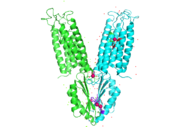
| - | YiiP has a "Y" shape <scene name='75/756372/Newmainpic/1'> conformation</scene> with two different functional conformations. An interlocked salt bridge connects the two domains with the Lys77 and the Asp207 from each | + | YiiP has a "Y" shape <scene name='75/756372/Newmainpic/1'> conformation</scene> with two different functional conformations. An interlocked salt bridge connects the two domains with the Lys77 and the Asp207 from each monomer. This [https://en.wikipedia.org/wiki/Salt_bridge_(protein_and_supramolecular) salt bridge] acts as the hinge for the conformational changes that YiiP undergoes. YiiP has three zinc binding sites, two of which are known to play an active role in the function of YiiP. |
===Interlocking Salt Bridge=== | ===Interlocking Salt Bridge=== | ||
Revision as of 21:30, 29 March 2017
Introduction
Zinc transporter is an integral membrane protein found in the membrane of Esherichia coli. YiiP is a member of the cation diffusion facilitator family. Members of this family occur all throughout the biological realm. These diffusion facilitators export divalent transition metal ions from the cytoplasm to the extracellular space [1]. They work to regulate the amount of divalent metals inside of the cell, which is biologically relevant because while these metals are necessary for different biological functions, they can prove fatal to the cell in excess amounts. Zinc is essential for the growth and development of cells and zinc levels can affect everything from gene expression to immune response. While YiiP is an integral membrane protein in the cells of Escherichia coli, understanding the mechanism of regulation behind it can help researcher's better understand the cation diffusion facilitator equivalents in eukaryotic cells.
| |||||||||||