User:Natalie Van Ochten/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
===Active Site=== | ===Active Site=== | ||
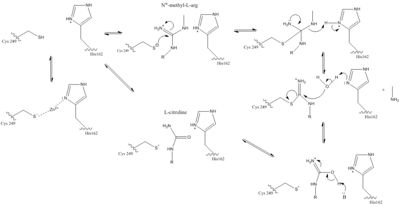
| - | The normal DDAH regulation <span class="plainlinks">[https://en.wikipedia.org/wiki/Reaction_mechanism mechanism]</span> depends on the presence of <scene name='75/752351/Ddah_active_site/2'>Cys 249</scene> in the active site that acts as a <span class="plainlinks">[https://en.wikipedia.org/wiki/Nucleophile nucleophile]</span> in the mechanism <ref name="stone">Stone EM, Costello AL, Tierney DL, Fast W. Substrate-assisted cysteine deprotonation in the mechanism of Dimethylargininase (DDAH) from Pseudomonas aeruginosa. Biochemistry. 2006 May 2;45(17):5618-5630. PMID:<span class="plainlinks">[https://www.ncbi.nlm.nih.gov/pubmed/16634643 16634643]</span> doi:<span class="plainlinks">[http://pubs.acs.org/doi/abs/10.1021/bi052595m 10.1021/bi052595m]</span></ref>. The Cys249 is used to attack the <span class="plainlinks">[https://en.wikipedia.org/wiki/Guanidine guanidinium]</span> carbon on the substrate that is held in the active site via hydrogen bonds. This is followed by collapsing the tetrahedral product to get rid of the <span class="plainlinks">[https://en.wikipedia.org/wiki/Alkylamines alkylamine]</span> leaving group. A <span class="plainlinks">[https://en.wikipedia.org/wiki/Isothiouronium thiouronium]</span> intermediate is then formed with <span class="plainlinks">[https://en.wikipedia.org/wiki/Orbital_hybridisation sp<sup>2</sup> hybridization]</span>. This intermediate is hydrolyzed to form citrulline. The His162 protonates the leaving group in this reaction and generates hydroxide to hydrolyze the intermediate formed in the reaction. Studies suggest that Cys249 is neutral until binding of guanidinium near Cys249 decreases Cys249’s <span class="plainlinks">[https://en.wikipedia.org/wiki/Acid_dissociation_constant pKa]</span> and deprotonates the thiolate to activate the nucleophile. Other studies suggest that the Cys249 and an active site His162 form an <span class="plainlinks">[https://en.wikipedia.org/wiki/Intimate_ion_pair ion pair]</span> to deprotonate the thiolate. Cys249 and His162 can also form a binding site for inhibitors to bind to which stabilizes the thiolate. This is important in regulating NO activity in organisms and designing drugs to inhibit this enzyme <ref name="stone" />. | + | The normal DDAH regulation <span class="plainlinks">[https://en.wikipedia.org/wiki/Reaction_mechanism mechanism]</span> depends on the presence of <scene name='75/752351/Ddah_active_site/2'>Cys 249</scene> in the active site that acts as a <span class="plainlinks">[https://en.wikipedia.org/wiki/Nucleophile nucleophile]</span> in the mechanism <ref name="stone">Stone EM, Costello AL, Tierney DL, Fast W. Substrate-assisted cysteine deprotonation in the mechanism of Dimethylargininase (DDAH) from Pseudomonas aeruginosa. Biochemistry. 2006 May 2;45(17):5618-5630. PMID:<span class="plainlinks">[https://www.ncbi.nlm.nih.gov/pubmed/16634643 16634643]</span> doi:<span class="plainlinks">[http://pubs.acs.org/doi/abs/10.1021/bi052595m 10.1021/bi052595m]</span></ref>. The Cys249 is used to attack the <span class="plainlinks">[https://en.wikipedia.org/wiki/Guanidine guanidinium]</span> carbon on the substrate that is held in the active site via hydrogen bonds (Figure 1). This is followed by collapsing the tetrahedral product to get rid of the <span class="plainlinks">[https://en.wikipedia.org/wiki/Alkylamines alkylamine]</span> leaving group. A <span class="plainlinks">[https://en.wikipedia.org/wiki/Isothiouronium thiouronium]</span> intermediate is then formed with <span class="plainlinks">[https://en.wikipedia.org/wiki/Orbital_hybridisation sp<sup>2</sup> hybridization]</span>. This intermediate is hydrolyzed to form citrulline. The His162 protonates the leaving group in this reaction and generates hydroxide to hydrolyze the intermediate formed in the reaction (Figure 1). Studies suggest that Cys249 is neutral until binding of guanidinium near Cys249 decreases Cys249’s <span class="plainlinks">[https://en.wikipedia.org/wiki/Acid_dissociation_constant pKa]</span> and deprotonates the thiolate to activate the nucleophile. Other studies suggest that the Cys249 and an active site His162 form an <span class="plainlinks">[https://en.wikipedia.org/wiki/Intimate_ion_pair ion pair]</span> to deprotonate the thiolate. Cys249 and His162 can also form a binding site for inhibitors to bind to which stabilizes the thiolate. This is important in regulating NO activity in organisms and designing drugs to inhibit this enzyme <ref name="stone" />. |
[[Image:The Normal DDAH Mechanism.jpg|400px|center|thumb|The normal DDAH mechanism]] | [[Image:The Normal DDAH Mechanism.jpg|400px|center|thumb|The normal DDAH mechanism]] | ||
Revision as of 16:49, 31 March 2017
Dimethylarginine Dimethylaminohydrolase
| |||||||||||
References
- ↑ 1.0 1.1 Palm F, Onozato ML, Luo Z, Wilcox CS. Dimethylarginine dimethylaminohydrolase (DDAH): expression, regulation, and function in the cardiovascular and renal systems. American Journal of Physiology. 2007 Dec 1;293(6):3227-3245. PMID:17933965 doi:10.1152/ajpheart.00998.2007
- ↑ 2.0 2.1 2.2 Tran CTL, Leiper JM, Vallance P. The DDAH/ADMA/NOS pathway. Atherosclerosis Supplements. 2003 Dec;4(4):33-40. PMID:14664901 doi:10.1016/S1567-5688(03)00032-1
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 3.11 3.12 3.13 3.14 3.15 3.16 3.17 3.18 3.19 3.20 3.21 3.22 3.23 Frey D, Braun O, Briand C, Vasak M, Grutter MG. Structure of the mammalian NOS regulator dimethylarginine dimethylaminohydrolase: a basis for the design of specific inhibitors. Structure. 2006 May;14(5):901-911. PMID:16698551 doi:10.1016/j.str.2006.03.006
- ↑ Janssen W, Pullamsetti SS, Cooke J, Weissmann N, Guenther A, Schermuly RT. The role of dimethylarginine dimethylaminohydrolase (DDAH) in pulmonary fibrosis. The Journal of Pathology. 2012 Dec 12;229(2):242-249. Epub 2013 Jan. PMID:23097221 doi:10.1002/path.4127
- ↑ Humm A, Fritsche E, Mann K, Göhl M, Huber R. Recombinant expression and isolation of human L-arginine:glycine amidinotransferase and identification of its active-site cysteine residue. Biochemical Journal. 1997 March 15;322(3):771-776. PMID:9148748 doi:10.1042/bj3220771
- ↑ 6.0 6.1 6.2 Rasheed M, Richter C, Chisty LT, Kirkpatrick J, Blackledge M, Webb MR, Driscoll PC. Ligand-dependent dynamics of the active site lid in bacterial Dimethyarginine Dimethylaminohydrolase. Biochemistry. 2014 Feb 18;53:1092-1104. PMCID:PMC3945819 doi:10.1021/bi4015924
- ↑ 7.0 7.1 Stone EM, Costello AL, Tierney DL, Fast W. Substrate-assisted cysteine deprotonation in the mechanism of Dimethylargininase (DDAH) from Pseudomonas aeruginosa. Biochemistry. 2006 May 2;45(17):5618-5630. PMID:16634643 doi:10.1021/bi052595m
- ↑ 8.0 8.1 Pace NJ, Weerpana E. Zinc-binding cysteines: diverse functions and structural motifs. Biomolecules. 2014 June;4(2):419-434. PMCID:4101490 doi:10.3390/biom4020419
- ↑ Colasanti M, Suzuki H. The dual personality of NO. ScienceDirect. 2000 Jul 1;21(7):249-252. PMID:10979862 doi:10.1016/S0165-6147(00)01499-1
- ↑ Rassaf T, Feelisch M, Kelm M. Circulating NO pool: assessment of nitrite and nitroso species in blood and tissues. Free Rad. Biol. Med. 2004 Feb 15;36(4):413-422. PMID:14975444 doi:10.1016/j.freeradbiomed.2003.11.011
- ↑ Tsao PS, Cooke JP. Endothelial alterations in hypercholesterolemia: more than simply vasodilator dysfunction. Journal of Cardiovascular Pharmacology. 1998;32(3):48-53. PMID:9883748
- ↑ Vallance P, Leiper J. Blocking NO synthesis: how, where and why? Nat. Rev. Drug Discov. 2002 Dec;1(12):939-950. PMID:12461516 doi:10.1038/nrd960
Student Contributors
- Natalie Van Ochten
- Kaitlyn Enderle
- Colton Junod