User:Natalie Van Ochten/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
===Lid Region=== | ===Lid Region=== | ||
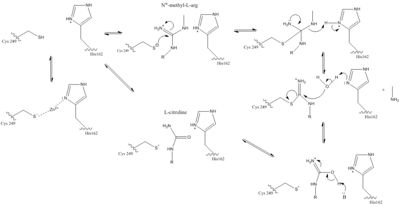
| - | Amino acids 25-36 of DDAH constitute the loop region of the protein <ref name="frey" />. This is more commonly known as the lid region. The lid is what allows the active site to be exposed to substrate binding or not. Studies have shown crystal structures of the lid at <scene name='69/694225/Low_ph_w_lid/1'>open</scene> and <scene name='69/694225/Closed_lid_zn9/1'>closed </scene> conformations. In the open conformation, the lid forms an alpha helix and the amino acid Leu29 is moved so it does not interact with the active site. This allows the active site to be vulnerable to attack. This lid region is very flexible. This open conformation has been shown when DDAH had been <span class="plainlinks">[https://en.wikipedia.org/wiki/Crystallization crystallized]</span> when <span class="plainlinks">[https://en.wikipedia.org/wiki/Zinc Zn(II)]</span> was bound at <scene name='69/694225/Active_site6/1'>pH 6.3</scene>. There is a closed form which has been observed with Zn(II) binding at <scene name='69/694225/Active_site_9/1'>pH 9.0</scene> and in the unliganded enzyme. When the lid is closed, a specific <span class="plainlinks">[https://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bond]</span> can form between the Leu29 carbonyl and the amino group on bound molecule. This stabilizes this complex. The Leu29 is then blocking the active site entrance <ref name="frey" />. Opening and closing the lid takes place faster than the actual reaction in the active site <ref name="rasheed">Rasheed M, Richter C, Chisty LT, Kirkpatrick J, Blackledge M, Webb MR, Driscoll PC. Ligand-dependent dynamics of the active site lid in bacterial Dimethyarginine Dimethylaminohydrolase. Biochemistry. 2014 Feb 18;53:1092-1104. PMCID:<span class="plainlinks">[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3945819/ PMC3945819]</span> doi:<span class="plainlinks">[http://pubs.acs.org/doi/abs/10.1021/bi4015924 10.1021/bi4015924]</span></ref>. This suggests that the <span class="plainlinks">[https://en.wikipedia.org/wiki/Rate-determining_step rate-limiting step]</span> of this reaction is not the lid movement but is the actual chemistry happening to the substrate in the active site of DDAH <ref name="rasheed" />. | + | Amino acids 25-36 of DDAH constitute the loop region of the protein <ref name="frey" />. This is more commonly known as the lid region. The lid is what allows the active site to be exposed to substrate binding or not. Studies have shown crystal structures of the lid at <scene name='69/694225/Low_ph_w_lid/1'>open</scene> and <scene name='69/694225/Closed_lid_zn9/1'>closed </scene> conformations. In the open conformation, the lid forms an alpha helix and the amino acid <scene name='69/694225/Low_ph_w_lid/2'>Leu29</scene> is moved so it does not interact with the active site. This allows the active site to be vulnerable to attack. This lid region is very flexible. This open conformation has been shown when DDAH had been <span class="plainlinks">[https://en.wikipedia.org/wiki/Crystallization crystallized]</span> when <span class="plainlinks">[https://en.wikipedia.org/wiki/Zinc Zn(II)]</span> was bound at <scene name='69/694225/Active_site6/1'>pH 6.3</scene>. There is a closed form which has been observed with Zn(II) binding at <scene name='69/694225/Active_site_9/1'>pH 9.0</scene> and in the unliganded enzyme. When the lid is closed, a specific <span class="plainlinks">[https://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bond]</span> can form between the Leu29 carbonyl and the amino group on bound molecule. This stabilizes this complex. The Leu29 is then blocking the active site entrance <ref name="frey" />. Opening and closing the lid takes place faster than the actual reaction in the active site <ref name="rasheed">Rasheed M, Richter C, Chisty LT, Kirkpatrick J, Blackledge M, Webb MR, Driscoll PC. Ligand-dependent dynamics of the active site lid in bacterial Dimethyarginine Dimethylaminohydrolase. Biochemistry. 2014 Feb 18;53:1092-1104. PMCID:<span class="plainlinks">[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3945819/ PMC3945819]</span> doi:<span class="plainlinks">[http://pubs.acs.org/doi/abs/10.1021/bi4015924 10.1021/bi4015924]</span></ref>. This suggests that the <span class="plainlinks">[https://en.wikipedia.org/wiki/Rate-determining_step rate-limiting step]</span> of this reaction is not the lid movement but is the actual chemistry happening to the substrate in the active site of DDAH <ref name="rasheed" />. |
The specific residues in the lid region are different in different organisms <ref name="frey" />. The only consistent similarity is a <span class="plainlinks">[https://en.wikipedia.org/wiki/Conserved_sequence conserved]</span> leucine residue in this lid that function to hydrogen bond with the <span class="plainlinks">[https://en.wikipedia.org/wiki/Ligand ligand]</span> bound to the active site <ref name="rasheed" />. Different <span class="plainlinks">[https://en.wikipedia.org/wiki/Protein_isoform isoforms]</span> from the same species can have differences in lid regions as well <ref name="frey" />. DDAH-2 has a negatively charged lid while DDAH-1 has a positively charged lid <ref name="frey" />. | The specific residues in the lid region are different in different organisms <ref name="frey" />. The only consistent similarity is a <span class="plainlinks">[https://en.wikipedia.org/wiki/Conserved_sequence conserved]</span> leucine residue in this lid that function to hydrogen bond with the <span class="plainlinks">[https://en.wikipedia.org/wiki/Ligand ligand]</span> bound to the active site <ref name="rasheed" />. Different <span class="plainlinks">[https://en.wikipedia.org/wiki/Protein_isoform isoforms]</span> from the same species can have differences in lid regions as well <ref name="frey" />. DDAH-2 has a negatively charged lid while DDAH-1 has a positively charged lid <ref name="frey" />. | ||
Revision as of 17:05, 31 March 2017
Dimethylarginine Dimethylaminohydrolase
| |||||||||||
References
- ↑ 1.0 1.1 Palm F, Onozato ML, Luo Z, Wilcox CS. Dimethylarginine dimethylaminohydrolase (DDAH): expression, regulation, and function in the cardiovascular and renal systems. American Journal of Physiology. 2007 Dec 1;293(6):3227-3245. PMID:17933965 doi:10.1152/ajpheart.00998.2007
- ↑ 2.0 2.1 2.2 Tran CTL, Leiper JM, Vallance P. The DDAH/ADMA/NOS pathway. Atherosclerosis Supplements. 2003 Dec;4(4):33-40. PMID:14664901 doi:10.1016/S1567-5688(03)00032-1
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 3.11 3.12 3.13 3.14 3.15 3.16 3.17 3.18 3.19 3.20 3.21 3.22 3.23 Frey D, Braun O, Briand C, Vasak M, Grutter MG. Structure of the mammalian NOS regulator dimethylarginine dimethylaminohydrolase: a basis for the design of specific inhibitors. Structure. 2006 May;14(5):901-911. PMID:16698551 doi:10.1016/j.str.2006.03.006
- ↑ Janssen W, Pullamsetti SS, Cooke J, Weissmann N, Guenther A, Schermuly RT. The role of dimethylarginine dimethylaminohydrolase (DDAH) in pulmonary fibrosis. The Journal of Pathology. 2012 Dec 12;229(2):242-249. Epub 2013 Jan. PMID:23097221 doi:10.1002/path.4127
- ↑ Humm A, Fritsche E, Mann K, Göhl M, Huber R. Recombinant expression and isolation of human L-arginine:glycine amidinotransferase and identification of its active-site cysteine residue. Biochemical Journal. 1997 March 15;322(3):771-776. PMID:9148748 doi:10.1042/bj3220771
- ↑ 6.0 6.1 6.2 Rasheed M, Richter C, Chisty LT, Kirkpatrick J, Blackledge M, Webb MR, Driscoll PC. Ligand-dependent dynamics of the active site lid in bacterial Dimethyarginine Dimethylaminohydrolase. Biochemistry. 2014 Feb 18;53:1092-1104. PMCID:PMC3945819 doi:10.1021/bi4015924
- ↑ 7.0 7.1 Stone EM, Costello AL, Tierney DL, Fast W. Substrate-assisted cysteine deprotonation in the mechanism of Dimethylargininase (DDAH) from Pseudomonas aeruginosa. Biochemistry. 2006 May 2;45(17):5618-5630. PMID:16634643 doi:10.1021/bi052595m
- ↑ 8.0 8.1 Pace NJ, Weerpana E. Zinc-binding cysteines: diverse functions and structural motifs. Biomolecules. 2014 June;4(2):419-434. PMCID:4101490 doi:10.3390/biom4020419
- ↑ Colasanti M, Suzuki H. The dual personality of NO. ScienceDirect. 2000 Jul 1;21(7):249-252. PMID:10979862 doi:10.1016/S0165-6147(00)01499-1
- ↑ Rassaf T, Feelisch M, Kelm M. Circulating NO pool: assessment of nitrite and nitroso species in blood and tissues. Free Rad. Biol. Med. 2004 Feb 15;36(4):413-422. PMID:14975444 doi:10.1016/j.freeradbiomed.2003.11.011
- ↑ Tsao PS, Cooke JP. Endothelial alterations in hypercholesterolemia: more than simply vasodilator dysfunction. Journal of Cardiovascular Pharmacology. 1998;32(3):48-53. PMID:9883748
- ↑ Vallance P, Leiper J. Blocking NO synthesis: how, where and why? Nat. Rev. Drug Discov. 2002 Dec;1(12):939-950. PMID:12461516 doi:10.1038/nrd960
Student Contributors
- Natalie Van Ochten
- Kaitlyn Enderle
- Colton Junod