Sandbox Reserved 1063
From Proteopedia
| Line 7: | Line 7: | ||
<StructureSection load='3TGN' size='350' side='right' caption='3TGN' scene=''> | <StructureSection load='3TGN' size='350' side='right' caption='3TGN' scene=''> | ||
== '''Zn(II) Binding''' == | == '''Zn(II) Binding''' == | ||
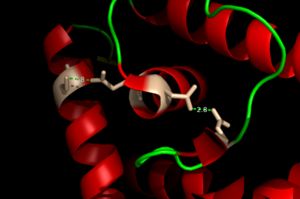
| - | Zinc-Dependent Transcriptional Regulator AdcR has <scene name='69/694230/Two_binding_sites/1'>two binding sites</scene> on each of its two protomers and can bind a total of four Zn(II) per dimer. <scene name='69/694230/Dimerization_domain/1'>The dimerization domain</scene> is made up of the <font color='blue'>alpha helix 1</font>, <font color='gold'>alpha helix 5</font>, and the <font color='orange'>alpha helix 6</font>. This domain is connected to the HTH winged domain with the long <font color='gold'>alpha helix 5</font>. This dimerization domain connects to the DNA binding domain and together with the <font color='blue'>alpha 1</font> <font color='turquoise'>alpha 2</font> loop make up the <scene name='69/694230/Alpha_1_alpha_2/1'>metal binding sites</scene><ref>PMID:22085181</ref>. Each protomer has one high affinity site (KZn1 = 10^12 M; pH 8) and one low affinity binding site (KZn2 = 10^7 M; pH 8). The metal binding pockets of the AdcR MarR transcriptional regulator are made up of the DNA binding domain with the extended alpha 1 and alpha 2 loop. The two different Zn(II) binding sites are connected via hydrogen bonding of the Nd1 atom of H108 and then liganding Oe1 atom of E41. | + | Zinc-Dependent Transcriptional Regulator AdcR has <scene name='69/694230/Two_binding_sites/1'>two binding sites</scene> on each of its two protomers and can bind a total of four Zn(II) per dimer. <scene name='69/694230/Dimerization_domain/1'>The dimerization domain</scene> is made up of the <font color='blue'>alpha helix 1</font>, <font color='gold'>alpha helix 5</font>, and the <font color='orange'>alpha helix 6</font>. This domain is connected to the HTH winged domain with the long <font color='gold'>alpha helix 5</font>. This dimerization domain connects to the DNA binding domain and together with the <font color='blue'>alpha 1</font> <font color='turquoise'>alpha 2</font> loop make up the <scene name='69/694230/Alpha_1_alpha_2/1'>metal binding sites</scene><ref>PMID:22085181</ref>. Each protomer has one high affinity site (KZn1 = 10^12 M; pH 8) and one low affinity binding site (KZn2 = 10^7 M; pH 8). The metal binding pockets of the AdcR MarR transcriptional regulator are made up of the DNA binding domain with the extended alpha 1 and alpha 2 loop. The two different Zn(II) binding sites are connected via <scene name='69/694230/Hydrogen_bonding/3'>hydrogen bonding</scene> of the Nd1 atom of H108 and then liganding Oe1 atom of E41. |
=== Binding Site 1 === | === Binding Site 1 === | ||
<scene name='69/694230/Binding_site_1/1'>Binding site 1</scene> consists of a distorted tetrahedral geometry around Zn(II). The four amino acids involved in zinc binding are E24 Oe1, H42 Nd1, H108 Ne2, and H112 Ne2. Binding site 1 is the only binding site that plays a significant role in the protein's regulatory function. The ability of binding site 1 to coordinate to the Zn(II) ion is pH dependent. At pH 6 the binding affinity for the Zn(II) ion is 10^9 - 10^10 M^-1, but at pH 8 the binding affinity increases to 10^12 M^-1. This is due to the charges on the histidines of the binding site. At pH 8, the histidines are positively charged and can interact with the negatively charged Zn(II) ion. However, at pH 6 the histidines are neutrally charged and will not coordinate as well with Zn(II). | <scene name='69/694230/Binding_site_1/1'>Binding site 1</scene> consists of a distorted tetrahedral geometry around Zn(II). The four amino acids involved in zinc binding are E24 Oe1, H42 Nd1, H108 Ne2, and H112 Ne2. Binding site 1 is the only binding site that plays a significant role in the protein's regulatory function. The ability of binding site 1 to coordinate to the Zn(II) ion is pH dependent. At pH 6 the binding affinity for the Zn(II) ion is 10^9 - 10^10 M^-1, but at pH 8 the binding affinity increases to 10^12 M^-1. This is due to the charges on the histidines of the binding site. At pH 8, the histidines are positively charged and can interact with the negatively charged Zn(II) ion. However, at pH 6 the histidines are neutrally charged and will not coordinate as well with Zn(II). | ||
Revision as of 18:09, 31 March 2017
Zinc-Dependent Transcriptional Regulator AdcR
Introduction
AdcR is a zinc-dependent transcriptional regulator that controls the activation of over seventy genes within the bacteria Streptococcus pneumoniae[1]. Zinc plays a vital role in organism homeostasis acting as a co-factor and a regulator of enzymatic activity, however zinc can lead to cell toxicity and deficiency of other vital metals that are also necessary for protein function[2][3]. Given the importance of zinc in general homeostasis the vital role of AdcR in Streptococcus pneumoniae can be understood given its ability to regulate zinc transfer proteins within the bacteria.
Consistent with AdcR's identity as a member of the MarR protein family, AdcR exhibits a triangular shape consisting of a two fold pseudosymetric homo dimer with its own unique winged helix-turn-helix (wHTH) binding domain. This structure calls for multiple zinc binding sites that facilitate protein conformational change allowing for DNA binding and regulation through the wHTH domain.
| |||||||||||
References
- ↑ Sanson M, Makthal N, Flores AR, Olsen RJ, Musser JM, Kumaraswami M. Adhesin competence repressor (AdcR) from Streptococcus pyogenes controls adaptive responses to zinc limitation and contributes to virulence. Nucleic Acids Res. 2015 Jan;43(1):418-32. doi: 10.1093/nar/gku1304. Epub 2014 Dec, 15. PMID:25510500 doi:http://dx.doi.org/10.1093/nar/gku1304
- ↑ Fraústo da Silva J, Williams R. The Biological Chemistry of Elements: The Inorganic Chemistry of Life. Second ed. Oxford University Press; Oxford: 2001.
- ↑ Ma Z, Jacobsen FE, Giedroc DP. Coordination chemistry of bacterial metal transport and sensing. Chem Rev. 2009 Oct;109(10):4644-81. doi: 10.1021/cr900077w. PMID:19788177 doi:http://dx.doi.org/10.1021/cr900077w
- ↑ Guerra AJ, Dann CE, Giedroc DP. Crystal Structure of the Zinc-Dependent MarR Family Transcriptional Regulator AdcR in the Zn(II)-Bound State. J Am Chem Soc. 2011 Nov 21. PMID:22085181 doi:10.1021/ja2080532