User:Luke Edward Severinac/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 10: | Line 10: | ||
===Active Site=== | ===Active Site=== | ||
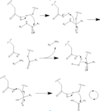
Caspase 6 is a part of the cystine aspartic protease family that cleaves proteins at the TEVD sequence. in its monomeric form with protein ligand bound, its catalytic residues are <scene name='75/752344/His121/1'>Histidine 121</scene>, <scene name='75/752344/Glu123/1'>Glutamate 123</scene>, and <scene name='75/752344/Cys163/1'>Cystine 163</scene>.[[Image:Mechanism caspase 6.PNG|100 px|left|thumb|Cystine Aspartase mechanism]] Together, these residues form a <scene name='75/752344/Caspase-6_catalytic_triad_real/1'>catalytic triad</scene> that cleaves a <scene name='75/752344/Protein_ligand/1'>protein ligand</scene>. | Caspase 6 is a part of the cystine aspartic protease family that cleaves proteins at the TEVD sequence. in its monomeric form with protein ligand bound, its catalytic residues are <scene name='75/752344/His121/1'>Histidine 121</scene>, <scene name='75/752344/Glu123/1'>Glutamate 123</scene>, and <scene name='75/752344/Cys163/1'>Cystine 163</scene>.[[Image:Mechanism caspase 6.PNG|100 px|left|thumb|Cystine Aspartase mechanism]] Together, these residues form a <scene name='75/752344/Caspase-6_catalytic_triad_real/1'>catalytic triad</scene> that cleaves a <scene name='75/752344/Protein_ligand/1'>protein ligand</scene>. | ||
| - | ===Zinc | + | ===Zinc Exosite=== |
=='''Activation of Caspase-6'''== | =='''Activation of Caspase-6'''== | ||
Caspase-6 has a small pro-domain. It shares 41% sequence identity with Caspase-3 and 37% sequence identity with Caspase-7 [[https://www.ncbi.nlm.nih.gov/pubmed/25340928]. Both of these caspases are classified as effectors and because of it's similarites to these other Caspases, Caspase-6 is also classified as an effector. Caspase-6, does however, have many unique features compared to the other effectors, it has similar substrate specificity to that of initiator Caspases-8 and -9. Inhibitors of Apoptosis or IAPs, which are known to inhibit Caspase-3, -7, and -9, do not inhibit Caspase-6. Caspase-6 is known to undergo self-processing and activation in vitro and in vivo. High activity of Caspase-6 protein do not induce apoptosis of in HEK293 cells. Caspase-6 also has a relatively low zymogenicty, which is the ratio of activity for cleaved protein to the activity of uncleaved protein, of about 200, which is comparable to the zymogenicities of Caspase-8 and Caspase-9, which are both classified as initiator caspases. Caspase-6’s zymogenicity is also much lower than Caspase-3, which is another effector. This is interesting because the Caspase-6 protein shows low activity when it is not cleaved, similar to the initiator caspases, but Caspase-3, another effector, has basically no zymogen activity. Caspase-6 is classified as an effector, but it can also act as an initiator and cleave Caspases-2 and -8. It can also induce the mitochondrial membrane to become permeable, which leads to cytochrome c release and activation of other effector caspases. | Caspase-6 has a small pro-domain. It shares 41% sequence identity with Caspase-3 and 37% sequence identity with Caspase-7 [[https://www.ncbi.nlm.nih.gov/pubmed/25340928]. Both of these caspases are classified as effectors and because of it's similarites to these other Caspases, Caspase-6 is also classified as an effector. Caspase-6, does however, have many unique features compared to the other effectors, it has similar substrate specificity to that of initiator Caspases-8 and -9. Inhibitors of Apoptosis or IAPs, which are known to inhibit Caspase-3, -7, and -9, do not inhibit Caspase-6. Caspase-6 is known to undergo self-processing and activation in vitro and in vivo. High activity of Caspase-6 protein do not induce apoptosis of in HEK293 cells. Caspase-6 also has a relatively low zymogenicty, which is the ratio of activity for cleaved protein to the activity of uncleaved protein, of about 200, which is comparable to the zymogenicities of Caspase-8 and Caspase-9, which are both classified as initiator caspases. Caspase-6’s zymogenicity is also much lower than Caspase-3, which is another effector. This is interesting because the Caspase-6 protein shows low activity when it is not cleaved, similar to the initiator caspases, but Caspase-3, another effector, has basically no zymogen activity. Caspase-6 is classified as an effector, but it can also act as an initiator and cleave Caspases-2 and -8. It can also induce the mitochondrial membrane to become permeable, which leads to cytochrome c release and activation of other effector caspases. | ||
| - | ===Structural Units involved=== | + | ===Structural Units involved in autoactivation=== |
It is expressed as a dimeric zymogen, it contains a short prodomain, a large subunuit, known as p20, an intersubunit linker, and a small subunit, known as p10. Caspase-6 contains three cleavage sites, the first following the residues TETD23 that follows the prodomain, the next sit follows the residue sequence DVVD179, and the third cleavage site falls within the intersubunit linker following the sequence TEVD193. To activate effector caspases there must be a cleavage at the intersubunit linker, which releases the N terminus of p10, the N terminus then rotates about 180⁰ to form a loop bundle with the four other loops of an adjacent catalytic unit, this stabilizes the substrate binding pockets. It is sufficient to cleave either or both of the intersubunit linkers to activate Caspase-6. It is also important to point out that the prodomain of Caspase-6 inhibits in vivo. Caspase-6 can either undergo autoactivation or it can be activated by Capase-3, but the patterns for these two modes of activation are different. When Caspase-6 is self-activating it loses the prodomain first by cleavage at TETD23. Then it self-cleaves at TEVD23, which results in the formation of the loop bundle. The final cleavage of autoactivation is at DVVD179. In comparison, when Caspase-3 is activating Caspase-6 the first cleavage is at DVVD179, then the next cleavage is at TETD23, and the final cleavage occurs at TEVD193. | It is expressed as a dimeric zymogen, it contains a short prodomain, a large subunuit, known as p20, an intersubunit linker, and a small subunit, known as p10. Caspase-6 contains three cleavage sites, the first following the residues TETD23 that follows the prodomain, the next sit follows the residue sequence DVVD179, and the third cleavage site falls within the intersubunit linker following the sequence TEVD193. To activate effector caspases there must be a cleavage at the intersubunit linker, which releases the N terminus of p10, the N terminus then rotates about 180⁰ to form a loop bundle with the four other loops of an adjacent catalytic unit, this stabilizes the substrate binding pockets. It is sufficient to cleave either or both of the intersubunit linkers to activate Caspase-6. It is also important to point out that the prodomain of Caspase-6 inhibits in vivo. Caspase-6 can either undergo autoactivation or it can be activated by Capase-3, but the patterns for these two modes of activation are different. When Caspase-6 is self-activating it loses the prodomain first by cleavage at TETD23. Then it self-cleaves at TEVD23, which results in the formation of the loop bundle. The final cleavage of autoactivation is at DVVD179. In comparison, when Caspase-3 is activating Caspase-6 the first cleavage is at DVVD179, then the next cleavage is at TETD23, and the final cleavage occurs at TEVD193. | ||
Revision as of 21:04, 2 April 2017
Caspase-6 in Homo sapiens
| |||||||||||
References
3. Wang, Xiao-Jun, Qin Cao, Yan Zhang, and Xiao-Dong Su. "Activation and Regulation of Caspase-6 and Its Role in Neurodegenerative Diseases." Annual Review of Pharmacology and Toxicology 55.1 (2015): 553-72. Web.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2966951/ (self cleavage article)
http://www.rcsb.org/pdb/explore/explore.do?structureId=2WDP (this is the non-self cleaved protien)