User:Luke Edward Severinac/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
=='''Structure'''== | =='''Structure'''== | ||
===Active Site=== | ===Active Site=== | ||
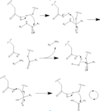
| - | In order to function as an endoprotease, Caspase-6 binds a <scene name='75/752344/Protein_ligand/1'>ligand</scene>, which can include neuronal proteins and tubulins [https://en.wikipedia.org/wiki/Tubulin], in its active site. This binding groove contains three critical amino acid residues necessary to perform cleavage of the peptide bonds. Together,<scene name='75/752344/His121/1'>His-121</scene>, <scene name='75/752344/Glu123/1'>Glu-123</scene>, and <scene name='75/752344/Cys163/1'>Cys-163</scene> form a <scene name='75/752344/Caspase-6_catalytic_triad_real/1'>catalytic triad</scene>[[Image:Mechanism caspase 6.PNG|100 px|left|thumb|Cystine Aspartase mechanism]]. In the theorized mechanism, His-121 provides hydrogen bonding to the carbonyl | + | In order to function as an endoprotease, Caspase-6 binds a <scene name='75/752344/Protein_ligand/1'>ligand</scene>, which can include neuronal proteins and tubulins [https://en.wikipedia.org/wiki/Tubulin], in its active site. This binding groove contains three critical amino acid residues necessary to perform cleavage of the peptide bonds. Together,<scene name='75/752344/His121/1'>His-121</scene>, <scene name='75/752344/Glu123/1'>Glu-123</scene>, and <scene name='75/752344/Cys163/1'>Cys-163</scene> form a <scene name='75/752344/Caspase-6_catalytic_triad_real/1'>catalytic triad</scene>[[Image:Mechanism caspase 6.PNG|100 px|left|thumb|Cystine Aspartase mechanism]]. In the theorized mechanism, His-121 provides hydrogen bonding to the carbonyl of the peptide ligand. This stabilizes the tetrahedral intermediate formed when Cys-163 nucleuphilically attacks the protein. Glu-123 is used in the catalytic triad to deprotonate Cys-163, increasing the nucleophilicity of the residue. |
===Zinc Exosite=== | ===Zinc Exosite=== | ||
| - | Caspase-6 function is inhibited by the binding of a zinc ion, which binds to an allosteric site instead of the active site. This allosteric site is located on the outside of the protein and it is distal to the active site. The Zinc ion is bound to three amino acid residues, Lysine-36, Glutamate-244, and Histidine-287, once the ion is bound to the protein it is then | + | Caspase-6 function is inhibited by the binding of a zinc ion, which binds to an allosteric site instead of the active site. This allosteric site is located on the outside of the protein and it is distal to the active site. The Zinc ion is bound to three amino acid residues, Lysine-36, Glutamate-244, and Histidine-287, once the ion is bound to the protein it is then stabilized by a single water molecule. The binding of Zinc at the exosite is suggested to cause a conformational change to the protein, which then causes a change in the active site, which inhibits Caspase-6's ability to bind to substrate. Zinc binding to the exosite is tightly regulated, because it inhibits Caspase-6's ability to inititate apoptosis. |
=='''Activation of Caspase-6'''== | =='''Activation of Caspase-6'''== | ||
Revision as of 22:51, 2 April 2017
Caspase-6 in Homo sapiens
| |||||||||||
References
Wang, Xiao-Jun, Qin Cao, Yan Zhang, and Xiao-Dong Su. "Activation and Regulation of Caspase-6 and Its Role in Neurodegenerative Diseases." Annual Review of Pharmacology and Toxicology 55.1 (2015): 553-72. Web.
Wang XJ, Cao Q, Liu X, Wang KT, Mi W, et al. 2010. Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation. EMBO Rep. 11: 841–47
(self cleavage article)
http://www.rcsb.org/pdb/explore/explore.do?structureId=2WDP (this is the non-self cleaved protien)