User:Luke Edward Severinac/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
<StructureSection load='4FXO' size='340' side='right' caption='Caspase-6' scene=''> | <StructureSection load='4FXO' size='340' side='right' caption='Caspase-6' scene=''> | ||
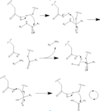
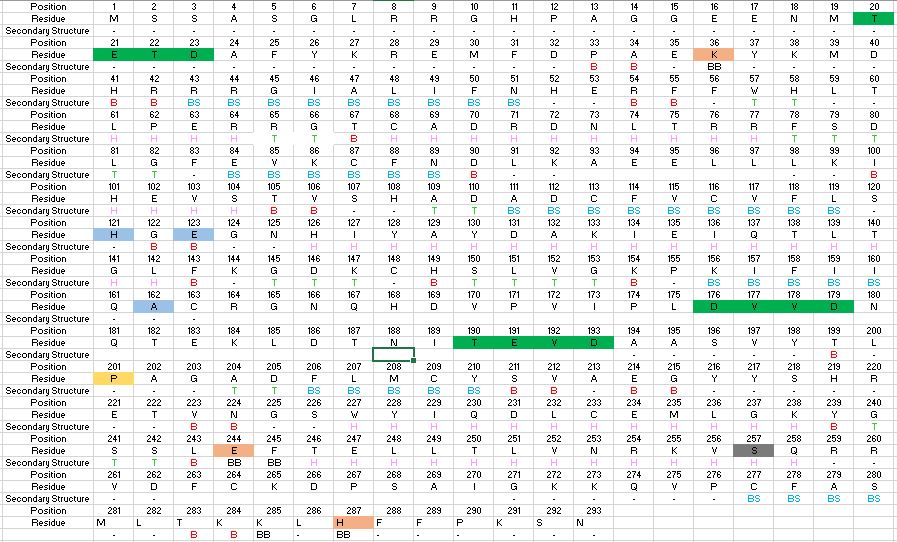
| - | Found at high concentrations in the brain and bordering tissues, Caspase-6 has been implicated in several neurological diseases including Alzheimer's and dementia[http://www.alz.org/]<ref name="ActiveRegofCasp6andNDdisease">PMID: 25340928 </ref>. It's primarily involved in apoptosis through a largely ambiguous mechanism. It is classified as an [https://en.wikipedia.org/wiki/Endopeptidase]endopeptidase as it cleaves an internal peptide bond of its substrate. It has relatively low specificity in the binding site which allows for a variety of substrates, including other caspase enzymes and neuronal proteins to bind<ref name="ZincMediatedCasp6">PMID: 22891250 </ref>. Furthermore, it is a part of the cysteine-aspartate family[https://en.wikipedia.org/wiki/Caspase], which have these critical amino acid residues in the active site of the enzyme. Caspase-6 has both an inactive zinc-bound conformation and an active ligand-bound conformation, which are largely regulated by variations in zinc concentration. | + | Found at high concentrations in the brain and bordering tissues, Caspase-6 has been implicated in several neurological diseases including Alzheimer's and dementia[http://www.alz.org/]<ref name="ActiveRegofCasp6andNDdisease">PMID: 25340928 </ref>. It's primarily involved in apoptosis through a largely ambiguous mechanism. It is classified as an [https://en.wikipedia.org/wiki/Endopeptidase]endopeptidase as it cleaves an internal peptide bond of its substrate. It has relatively low specificity in the binding site which allows for a variety of substrates, including other caspase enzymes and neuronal proteins to bind<ref name="ZincMediatedCasp6">PMID: 22891250 </ref>. Furthermore, it is a part of the cysteine-aspartate family[https://en.wikipedia.org/wiki/Caspase], which have these critical amino acid residues in the active site of the enzyme. Caspase-6 has both an inactive zinc-bound conformation and an active ligand-bound conformation, which are largely regulated by variations in zinc concentration<ref name="ZincMediatedCasp6">PMID: 22891250 </ref>. |
[[Image:Caspase-6 protein.jpg|100 px|left|thumb|Figure Legend]] | [[Image:Caspase-6 protein.jpg|100 px|left|thumb|Figure Legend]] | ||
Revision as of 23:42, 2 April 2017
Caspase-6 in Homo sapiens
| |||||||||||
References
- ↑ Wang XJ, Cao Q, Zhang Y, Su XD. Activation and regulation of caspase-6 and its role in neurodegenerative diseases. Annu Rev Pharmacol Toxicol. 2015;55:553-72. doi:, 10.1146/annurev-pharmtox-010814-124414. Epub 2014 Oct 17. PMID:25340928 doi:http://dx.doi.org/10.1146/annurev-pharmtox-010814-124414
- ↑ 2.0 2.1 Velazquez-Delgado EM, Hardy JA. Zinc-Mediated Allosteric Inhibition of Caspase-6. J Biol Chem. 2012 Aug 13. PMID:22891250 doi:http://dx.doi.org/10.1074/jbc.M112.397752
Wang, Xiao-Jun, Qin Cao, Yan Zhang, and Xiao-Dong Su. "Activation and Regulation of Caspase-6 and Its Role in Neurodegenerative Diseases." Annual Review of Pharmacology and Toxicology 55.1 (2015): 553-72. Web.
Wang XJ, Cao Q, Liu X, Wang KT, Mi W, et al. 2010. Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation. EMBO Rep. 11: 841–47
(self cleavage article)
http://www.rcsb.org/pdb/explore/explore.do?structureId=2WDP (this is the non-self cleaved protien)