User:Luke Edward Severinac/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 12: | Line 12: | ||
===Zinc Exosite=== | ===Zinc Exosite=== | ||
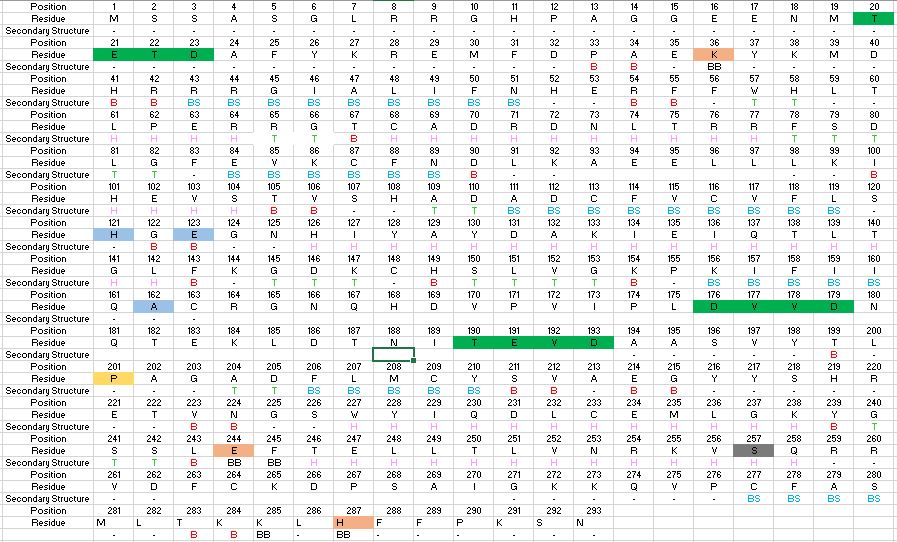
| - | Caspase-6 function is inhibited by the binding of a <scene name='75/752344/Zinc_caspase-6/1'>Zinc</scene> ion, which binds to an <scene name='75/752344/Caspase6_allosteric_site/1'>allosteric site</scene> instead of the <scene name='75/752344/Caspase6_allostericactiv_site/1'>active site</scene>. This allosteric site is located on the outside of the protein and it is distal to the active site. The Zinc ion is bound to <scene name='75/752344/Caspase6_allostericactiv_site/1'>three residues</scene>, Lys-36, Glu-244, and His-287, once the ion is bound to the protein it is then stabilized by a | + | Caspase-6 function is inhibited by the binding of a <scene name='75/752344/Zinc_caspase-6/1'>Zinc</scene> ion, which binds to an <scene name='75/752344/Caspase6_allosteric_site/1'>allosteric site</scene> instead of the <scene name='75/752344/Caspase6_allostericactiv_site/1'>active site</scene>. This allosteric site is located on the outside of the protein and it is distal to the active site. The Zinc ion is bound to <scene name='75/752344/Caspase6_allostericactiv_site/1'>three residues</scene>, Lys-36, Glu-244, and His-287, once the ion is bound to the protein it is then stabilized by a single water molecule. The binding of Zinc at the exosite is suggested to cause a conformational change to the protein, which then causes a change in the active site, which inhibits Caspase-6's ability to bind to substrate. Zinc binding to the exosite is tightly regulated, because it inhibits Caspase-6's ability to inititate apoptosis. |
=='''Activation of Caspase-6'''== | =='''Activation of Caspase-6'''== | ||
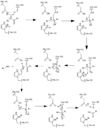
| - | Before Caspase-6 is a functional and active dimer, the enzyme exists as a procaspase, also known as zymogen [https://en.wikipedia.org/wiki/Zymogen]. Caspase-6 can be activated by acting as a substrate for other caspases, particularly Caspase-3, and enzymes. It becomes cleaved by these enzymes and proceeds to its active dimer conformation. When it was observed that Caspase-6 became active without other enzymes present, it was suggested that Caspase-6 utilizes a self-cleavage mechanism. Now, self-processing, a characteristic unique to Caspase-6, is recognized as the primary mechanism for Caspase-6 activation. The unprocessed enzyme contains a small and large subunit, a pro-domain, as well as an intersubunit linker. To become active, the intersubunit linker binds to the active site, where it is then cleaved. Other cleavages must occur as well for the enzyme to become active, specifically at TETD23 of the pro-domain, DVVD179, and TEVD193 amino acid sequences. Cleavage at these sites occurs in a specific sequence. To begin, the site within the pro-domain, TETD23, must be cleaved. This cleavage is then followed by either DVVD179 or TEVD193. | + | Before Caspase-6 is a functional and active dimer, the enzyme exists as a procaspase, also known as zymogen [https://en.wikipedia.org/wiki/Zymogen]. Caspase-6 can be activated by acting as a substrate for other caspases, particularly Caspase-3, and enzymes. It becomes cleaved by these enzymes and proceeds to its active dimer conformation. When it was observed that Caspase-6 became active without other enzymes present, it was suggested that Caspase-6 utilizes a self-cleavage mechanism. Now, self-processing, a characteristic unique to Caspase-6, is recognized as the primary mechanism for Caspase-6 activation. The unprocessed enzyme contains a small and large subunit, a pro-domain, as well as an intersubunit linker. To become active, the intersubunit linker binds to the active site, where it is then cleaved. Other cleavages must occur as well for the enzyme to become active, specifically at TETD23 of the pro-domain, DVVD179, and TEVD193 amino acid sequences<ref name="CrystalStrofhumCasp6revnewmech">PMID:20890311</ref>. Cleavage at these sites occurs in a specific sequence. To begin, the site within the pro-domain, TETD23, must be cleaved. This cleavage is then followed by either DVVD179 or TEVD193<ref name="ActiveRegofCasp6andNDdisease">PMID: 25340928 </ref>. |
| - | Despite the sequence similarities between TETD23 and TEVD193 cleavage sites, the TETD23 cleavage site is always cleaved before TEVD193. It has been proposed that this sequence of cleavage is due to the structure of Caspase-6, which allows the pro-domain to be more readily available to enter the active site. To some extent, the pro-domain inhibits Caspase-6's ability to cleave the intersubunit loop and self-activate, but this happens in a currently unknown mechanism. The result of the TETD23 cleavage site priority is that the prodomain acts as a “suicide protector”, which protects the TEVD193 cleavage site from self-cleavage[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2966951/]. This protection is necessary when there are low levels of proteins, which must be preserved, in the tissue. The pro-domain is released after the cleavage at TETD23 and cleavage of the intersubunit links follow. then allows the two subunits to interact to form the active dimer. The intramolecular cleavage of TEVD193 is essential for the initiation caspase-6 activation without other caspases, Caspase-3 in particular, present. | + | Despite the sequence similarities between TETD23 and TEVD193 cleavage sites, the TETD23 cleavage site is always cleaved before TEVD193. It has been proposed that this sequence of cleavage is due to the structure of Caspase-6, which allows the pro-domain to be more readily available to enter the active site. To some extent, the pro-domain inhibits Caspase-6's ability to cleave the intersubunit loop and self-activate, but this happens in a currently unknown mechanism. The result of the TETD23 cleavage site priority is that the prodomain acts as a “suicide protector”, which protects the TEVD193 cleavage site from self-cleavage[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2966951/]<ref name="ActiveRegofCasp6andNDdisease">PMID: 25340928 </ref>. This protection is necessary when there are low levels of proteins, which must be preserved, in the tissue. The pro-domain is released after the cleavage at TETD23 and cleavage of the intersubunit links follow. then allows the two subunits to interact to form the active dimer. The intramolecular cleavage of TEVD193 is essential for the initiation caspase-6 activation without other caspases, Caspase-3 in particular, present. |
Revision as of 03:22, 3 April 2017
Caspase-6 in Homo sapiens
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Wang XJ, Cao Q, Zhang Y, Su XD. Activation and regulation of caspase-6 and its role in neurodegenerative diseases. Annu Rev Pharmacol Toxicol. 2015;55:553-72. doi:, 10.1146/annurev-pharmtox-010814-124414. Epub 2014 Oct 17. PMID:25340928 doi:http://dx.doi.org/10.1146/annurev-pharmtox-010814-124414
- ↑ 2.0 2.1 Velazquez-Delgado EM, Hardy JA. Zinc-Mediated Allosteric Inhibition of Caspase-6. J Biol Chem. 2012 Aug 13. PMID:22891250 doi:http://dx.doi.org/10.1074/jbc.M112.397752
- ↑ Wang XJ, Cao Q, Liu X, Wang KT, Mi W, Zhang Y, Li LF, Leblanc AC, Su XD. Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation. EMBO Rep. 2010 Oct 1. PMID:20890311 doi:10.1038/embor.2010.141
Wang, Xiao-Jun, Qin Cao, Yan Zhang, and Xiao-Dong Su. "Activation and Regulation of Caspase-6 and Its Role in Neurodegenerative Diseases." Annual Review of Pharmacology and Toxicology 55.1 (2015): 553-72. Web.
Wang XJ, Cao Q, Liu X, Wang KT, Mi W, et al. 2010. Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation. EMBO Rep. 11: 841–47
(self cleavage article)
http://www.rcsb.org/pdb/explore/explore.do?structureId=2WDP (this is the non-self cleaved protien)