User:Luke Edward Severinac/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
<StructureSection load='4FXO' size='340' side='right' caption='Caspase-6' scene=''> | <StructureSection load='4FXO' size='340' side='right' caption='Caspase-6' scene=''> | ||
| - | Found at high concentrations in the brain and bordering tissues, Caspase-6 has been implicated in several neurological diseases including Alzheimer's and dementia[http://www.alz.org/]<ref name="ActiveRegofCasp6andNDdisease">PMID: 25340928 </ref>. It's primarily involved in apoptosis through a largely ambiguous mechanism. It is classified as an [https://en.wikipedia.org/wiki/Endopeptidase] | + | Found at high concentrations in the brain and bordering tissues, Caspase-6 has been implicated in several neurological diseases including Alzheimer's and dementia[http://www.alz.org/]<ref name="ActiveRegofCasp6andNDdisease">PMID: 25340928 </ref>. It's primarily involved in apoptosis through a largely ambiguous mechanism. It is classified as an endopeptidase[https://en.wikipedia.org/wiki/Endopeptidase] as it cleaves an internal peptide bond of its substrate. It has relatively low specificity in the binding site which allows for a variety of substrates, including other caspase enzymes and neuronal proteins to bind<ref name="ZincMediatedCasp6">PMID: 22891250 </ref>. Furthermore, it is a part of the cysteine-aspartate family[https://en.wikipedia.org/wiki/Caspase], which have these critical amino acid residues in the active site of the enzyme. Caspase-6 has both an inactive zinc-bound conformation and an active ligand-bound conformation, which are largely regulated by variations in zinc concentration<ref name="ZincMediatedCasp6">PMID: 22891250 </ref>. |
[[Image:Caspase-6 protein.jpg|100 px|left|thumb|Figure Legend]] | [[Image:Caspase-6 protein.jpg|100 px|left|thumb|Figure Legend]] | ||
| Line 8: | Line 8: | ||
[[Image:4FXO.PNG|100 px|left|thumb|This is the figure legend of the thumbnail]] | [[Image:4FXO.PNG|100 px|left|thumb|This is the figure legend of the thumbnail]] | ||
=='''Sequence and Structure'''== | =='''Sequence and Structure'''== | ||
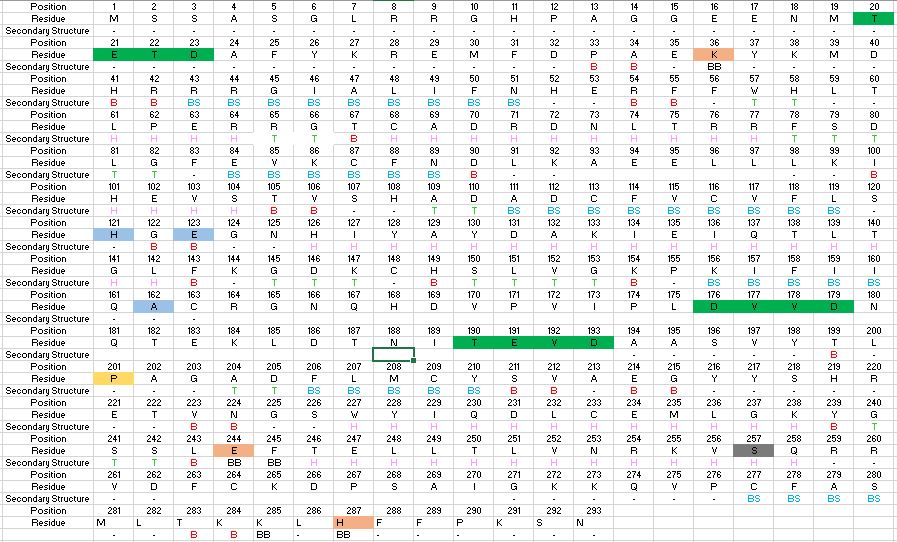
| - | The image below describes the amino acid sequence of Caspase-6, highlighting critical amino acids and sequences necessary for function | + | The image below describes the amino acid sequence of Caspase-6, highlighting critical amino acids and sequences necessary for function. It also highlights the secondary structures, which make up the folded protein, and sequences which become cleaved during zymogen processing. |
[[Image:Caspase 6 sequence image.JPG]][[Image:Caspase 6 sequence image key.JPG]] | [[Image:Caspase 6 sequence image.JPG]][[Image:Caspase 6 sequence image key.JPG]] | ||
| Line 29: | Line 29: | ||
===Phosphorylation=== | ===Phosphorylation=== | ||
| - | The function of Caspase-6 can be inhibited by phosphorylation of Ser-257. The exact mechanism of this reaction remains unidentified at the time of publication, but proceeds when ARK5 kinase is present. This modification can occur before and after zymogen activation or auto-processing. The phosphoryl group inhibits Caspase-6 through steric interference. When Ser-257 is phosphorylated, the amino acid residue interacts with <scene name='75/752344/Caspase-6_his-208/1'>Pro-201</scene>, causing a shift in the helices of Caspase-6. This is shown in the <scene name='75/752344/Caspase-6_s257d_mutant/1'>S257D Caspase-6 mutant</scene> mutant, whose mutation mimics phosphorylation. | + | The function of Caspase-6 can be inhibited by phosphorylation of Ser-257. The exact mechanism of this reaction remains unidentified at the time of publication, but proceeds when ARK5 kinase is present. This modification can occur before and after zymogen activation or auto-processing. The phosphoryl group inhibits Caspase-6 through steric interference. When Ser-257 is phosphorylated, the amino acid residue interacts with <scene name='75/752344/Caspase-6_his-208/1'>Pro-201</scene>, causing a shift in the helices of Caspase-6. This is shown in the <scene name='75/752344/Caspase-6_s257d_mutant/1'>S257D Caspase-6 mutant</scene> mutant, whose mutation mimics phosphorylation. <ref name="Phosregcasp6subsbindgroove">PMID: 22483120 </ref> The shift misaligns and disrupts residues found in the active site. This conformational difference prevents the inter-subunit loop from entering during zymogen activation and the self-cleaved active dimer cannot be formed. Additionally, no new substrate is able to enter the active site. |
Revision as of 00:31, 4 April 2017
Caspase-6 in Homo sapiens
| |||||||||||
References
- ↑ Wang XJ, Cao Q, Zhang Y, Su XD. Activation and regulation of caspase-6 and its role in neurodegenerative diseases. Annu Rev Pharmacol Toxicol. 2015;55:553-72. doi:, 10.1146/annurev-pharmtox-010814-124414. Epub 2014 Oct 17. PMID:25340928 doi:http://dx.doi.org/10.1146/annurev-pharmtox-010814-124414
- ↑ 2.0 2.1 Velazquez-Delgado EM, Hardy JA. Zinc-Mediated Allosteric Inhibition of Caspase-6. J Biol Chem. 2012 Aug 13. PMID:22891250 doi:http://dx.doi.org/10.1074/jbc.M112.397752
- ↑ Velazquez-Delgado EM, Hardy JA. Phosphorylation regulates assembly of the caspase-6 substrate-binding groove. Structure. 2012 Apr 4;20(4):742-51. Epub 2012 Apr 3. PMID:22483120 doi:10.1016/j.str.2012.02.003
Wang, Xiao-Jun, Qin Cao, Yan Zhang, and Xiao-Dong Su. "Activation and Regulation of Caspase-6 and Its Role in Neurodegenerative Diseases." Annual Review of Pharmacology and Toxicology 55.1 (2015): 553-72. Web.
Wang XJ, Cao Q, Liu X, Wang KT, Mi W, et al. 2010. Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation. EMBO Rep. 11: 841–47
(self cleavage article)
http://www.rcsb.org/pdb/explore/explore.do?structureId=2WDP (this is the non-self cleaved protien)