User:Natalie Van Ochten/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
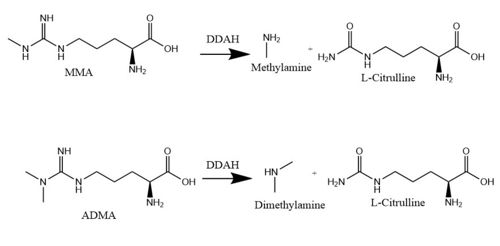
Dimethylarginine Dimethyaminohydrolase <span class="plainlinks">[http://www.chem.qmul.ac.uk/iubmb/enzyme/EC3/5/3/18.html EC 3.5.3.18]</span> (commonly known as DDAH) is a member of the <span class="plainlinks">[https://en.wikipedia.org/wiki/Hydrolase hydrolase]</span> family of enzymes which use water to break down molecules <ref name="palm">Palm F, Onozato ML, Luo Z, Wilcox CS. Dimethylarginine dimethylaminohydrolase (DDAH): expression, regulation, and function in the cardiovascular and renal systems. American Journal of Physiology. 2007 Dec 1;293(6):3227-3245. PMID:<span class="plainlinks">[https://www.ncbi.nlm.nih.gov/pubmed/17933965 17933965]</span> doi:<span class="plainlinks">[http://ajpheart.physiology.org/content/293/6/H3227 10.1152/ajpheart.00998.2007]</span></ref>. Additionally, DDAH is a <span class="plainlinks">[https://en.wikipedia.org/wiki/Nitric_oxide_synthase nitric oxide synthase (NOS)]</span> regulator. It metabolizes free arginine derivatives, namely <span class="plainlinks">[https://en.wikipedia.org/wiki/Asymmetric_dimethylarginine N<sup>Ѡ</sup>,N<sup>Ѡ</sup>-dimethyl-L-arginine (ADMA)]</span> and <span class="plainlinks">[https://en.wikipedia.org/wiki/Methylarginine NѠ-methyl-L-arginine (MMA)]</span> which competitively inhibit NOS <ref name="tran">Tran CTL, Leiper JM, Vallance P. The DDAH/ADMA/NOS pathway. Atherosclerosis Supplements. 2003 Dec;4(4):33-40. PMID:<span class="plainlinks">[https://www.ncbi.nlm.nih.gov/pubmed/14664901 14664901]</span> doi:<span class="plainlinks">[http://www.sciencedirect.com/science/article/pii/S1567568803000321 10.1016/S1567-5688(03)00032-1]</span></ref>. DDAH converts MMA and ADMA to <span class="plainlinks">[https://en.wikipedia.org/wiki/Citrulline L-citrulline]</span> and monoamine or dimethylamine <ref name="frey">Frey D, Braun O, Briand C, Vasak M, Grutter MG. Structure of the mammalian NOS regulator dimethylarginine dimethylaminohydrolase: a basis for the design of specific inhibitors. Structure. 2006 May;14(5):901-911. PMID:<span class="plainlinks">[https://www.ncbi.nlm.nih.gov/pubmed/16698551 16698551]</span> doi:<span class="plainlinks">[http://www.sciencedirect.com/science/article/pii/S0969212606001717 10.1016/j.str.2006.03.006]</span></ref>. DDAH is expressed in the cytosol of cells in humans, mice, rates, sheep, cattle, and bacteria <ref name="palm" />. DDAH activity has been localized mainly to the brain, kidney, pancreas, and liver in these organisms. Presented in this page is information from DDAH isoform 1; however, there are two different isoforms <ref name="frey" />. | Dimethylarginine Dimethyaminohydrolase <span class="plainlinks">[http://www.chem.qmul.ac.uk/iubmb/enzyme/EC3/5/3/18.html EC 3.5.3.18]</span> (commonly known as DDAH) is a member of the <span class="plainlinks">[https://en.wikipedia.org/wiki/Hydrolase hydrolase]</span> family of enzymes which use water to break down molecules <ref name="palm">Palm F, Onozato ML, Luo Z, Wilcox CS. Dimethylarginine dimethylaminohydrolase (DDAH): expression, regulation, and function in the cardiovascular and renal systems. American Journal of Physiology. 2007 Dec 1;293(6):3227-3245. PMID:<span class="plainlinks">[https://www.ncbi.nlm.nih.gov/pubmed/17933965 17933965]</span> doi:<span class="plainlinks">[http://ajpheart.physiology.org/content/293/6/H3227 10.1152/ajpheart.00998.2007]</span></ref>. Additionally, DDAH is a <span class="plainlinks">[https://en.wikipedia.org/wiki/Nitric_oxide_synthase nitric oxide synthase (NOS)]</span> regulator. It metabolizes free arginine derivatives, namely <span class="plainlinks">[https://en.wikipedia.org/wiki/Asymmetric_dimethylarginine N<sup>Ѡ</sup>,N<sup>Ѡ</sup>-dimethyl-L-arginine (ADMA)]</span> and <span class="plainlinks">[https://en.wikipedia.org/wiki/Methylarginine NѠ-methyl-L-arginine (MMA)]</span> which competitively inhibit NOS <ref name="tran">Tran CTL, Leiper JM, Vallance P. The DDAH/ADMA/NOS pathway. Atherosclerosis Supplements. 2003 Dec;4(4):33-40. PMID:<span class="plainlinks">[https://www.ncbi.nlm.nih.gov/pubmed/14664901 14664901]</span> doi:<span class="plainlinks">[http://www.sciencedirect.com/science/article/pii/S1567568803000321 10.1016/S1567-5688(03)00032-1]</span></ref>. DDAH converts MMA and ADMA to <span class="plainlinks">[https://en.wikipedia.org/wiki/Citrulline L-citrulline]</span> and monoamine or dimethylamine <ref name="frey">Frey D, Braun O, Briand C, Vasak M, Grutter MG. Structure of the mammalian NOS regulator dimethylarginine dimethylaminohydrolase: a basis for the design of specific inhibitors. Structure. 2006 May;14(5):901-911. PMID:<span class="plainlinks">[https://www.ncbi.nlm.nih.gov/pubmed/16698551 16698551]</span> doi:<span class="plainlinks">[http://www.sciencedirect.com/science/article/pii/S0969212606001717 10.1016/j.str.2006.03.006]</span></ref>. DDAH is expressed in the cytosol of cells in humans, mice, rates, sheep, cattle, and bacteria <ref name="palm" />. DDAH activity has been localized mainly to the brain, kidney, pancreas, and liver in these organisms. Presented in this page is information from DDAH isoform 1; however, there are two different isoforms <ref name="frey" />. | ||
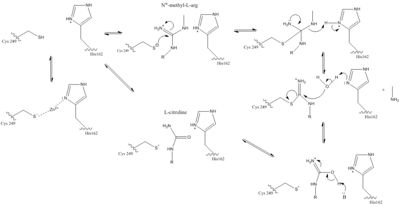
| - | [[Image:DDAH mechanism.jpg|500 px|center|thumb|Figure 1.]] | + | [[Image: DDAH mechanism.jpg|500 px|center|thumb|Figure 1.]] |
==Different Isoforms== | ==Different Isoforms== | ||
Revision as of 13:59, 11 April 2017
Dimethylarginine Dimethylaminohydrolase
| |||||||||||
References
- ↑ 1.0 1.1 Palm F, Onozato ML, Luo Z, Wilcox CS. Dimethylarginine dimethylaminohydrolase (DDAH): expression, regulation, and function in the cardiovascular and renal systems. American Journal of Physiology. 2007 Dec 1;293(6):3227-3245. PMID:17933965 doi:10.1152/ajpheart.00998.2007
- ↑ 2.0 2.1 2.2 Tran CTL, Leiper JM, Vallance P. The DDAH/ADMA/NOS pathway. Atherosclerosis Supplements. 2003 Dec;4(4):33-40. PMID:14664901 doi:10.1016/S1567-5688(03)00032-1
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 3.11 3.12 3.13 3.14 3.15 3.16 3.17 3.18 3.19 3.20 3.21 3.22 3.23 Frey D, Braun O, Briand C, Vasak M, Grutter MG. Structure of the mammalian NOS regulator dimethylarginine dimethylaminohydrolase: a basis for the design of specific inhibitors. Structure. 2006 May;14(5):901-911. PMID:16698551 doi:10.1016/j.str.2006.03.006
- ↑ Humm A, Fritsche E, Mann K, Göhl M, Huber R. Recombinant expression and isolation of human L-arginine:glycine amidinotransferase and identification of its active-site cysteine residue. Biochemical Journal. 1997 March 15;322(3):771-776. PMID:9148748 doi:10.1042/bj3220771
- ↑ 5.0 5.1 5.2 Rasheed M, Richter C, Chisty LT, Kirkpatrick J, Blackledge M, Webb MR, Driscoll PC. Ligand-dependent dynamics of the active site lid in bacterial Dimethyarginine Dimethylaminohydrolase. Biochemistry. 2014 Feb 18;53:1092-1104. PMCID:PMC3945819 doi:10.1021/bi4015924
- ↑ 6.0 6.1 Stone EM, Costello AL, Tierney DL, Fast W. Substrate-assisted cysteine deprotonation in the mechanism of Dimethylargininase (DDAH) from Pseudomonas aeruginosa. Biochemistry. 2006 May 2;45(17):5618-5630. PMID:16634643 doi:10.1021/bi052595m
- ↑ 7.0 7.1 Pace NJ, Weerpana E. Zinc-binding cysteines: diverse functions and structural motifs. Biomolecules. 2014 June;4(2):419-434. PMCID:4101490 doi:10.3390/biom4020419
- ↑ Janssen W, Pullamsetti SS, Cooke J, Weissmann N, Guenther A, Schermuly RT. The role of dimethylarginine dimethylaminohydrolase (DDAH) in pulmonary fibrosis. The Journal of Pathology. 2012 Dec 12;229(2):242-249. Epub 2013 Jan. PMID:23097221 doi:10.1002/path.4127
- ↑ Colasanti M, Suzuki H. The dual personality of NO. ScienceDirect. 2000 Jul 1;21(7):249-252. PMID:10979862 doi:10.1016/S0165-6147(00)01499-1
- ↑ Rassaf T, Feelisch M, Kelm M. Circulating NO pool: assessment of nitrite and nitroso species in blood and tissues. Free Rad. Biol. Med. 2004 Feb 15;36(4):413-422. PMID:14975444 doi:10.1016/j.freeradbiomed.2003.11.011
- ↑ Tsao PS, Cooke JP. Endothelial alterations in hypercholesterolemia: more than simply vasodilator dysfunction. Journal of Cardiovascular Pharmacology. 1998;32(3):48-53. PMID:9883748
- ↑ Vallance P, Leiper J. Blocking NO synthesis: how, where and why? Nat. Rev. Drug Discov. 2002 Dec;1(12):939-950. PMID:12461516 doi:10.1038/nrd960
Student Contributors
- Natalie Van Ochten
- Kaitlyn Enderle
- Colton Junod
3D Structures of Dimethylarginine Dimethylaminohydrolase
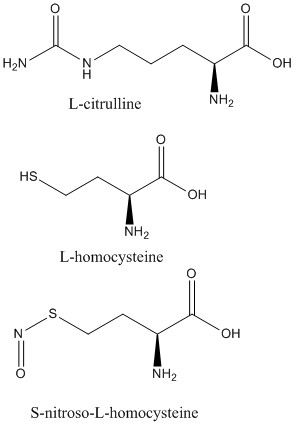
2C6Z (L-citrulline bound to isoform 1)
2CI1 (S-nitroso-L-homocysteine bound to isoform 1)
2CI3 (crystal form 1)
2CI4 (crystal form 2)
2CI5 (L-homocysteine bound to isoform 1)
2CI6 (Zn (II) bound at low pH to isoform 1)
2CI7 (Zn (II) bound at high pH to isoform 1)