User:Luke Edward Severinac/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
<StructureSection load='4FXO' size='340' side='right' caption='Caspase-6' scene=''> | <StructureSection load='4FXO' size='340' side='right' caption='Caspase-6' scene=''> | ||
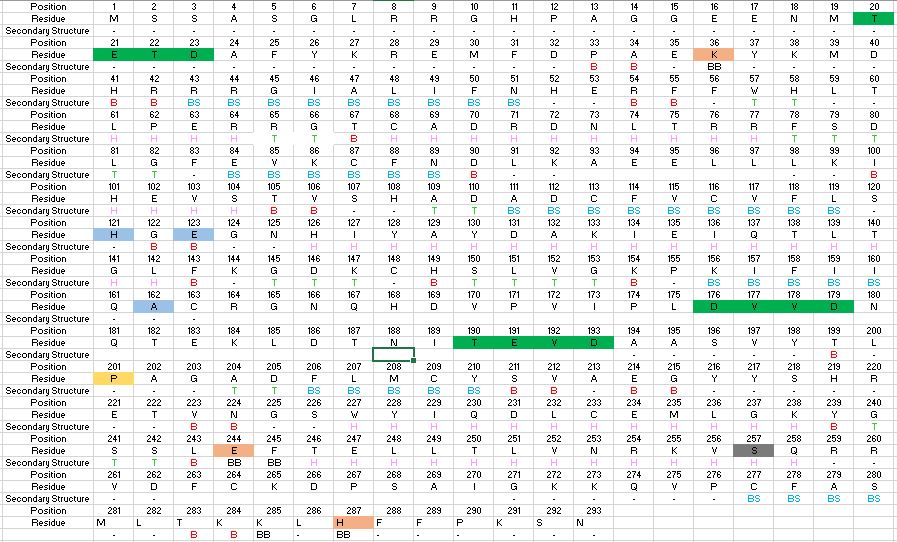
Found at high concentrations in the brain and bordering tissues, Caspase-6 has been implicated in several neurological diseases including Alzheimer's and dementia[http://www.alz.org/]<ref name="ActiveRegofCasp6andNDdisease">PMID: 25340928 </ref>. It's primarily involved in apoptosis through a largely ambiguous mechanism. It is classified as an endopeptidase[https://en.wikipedia.org/wiki/Endopeptidase] as it cleaves an internal peptide bond of its substrate. It has relatively low specificity in the binding site which allows for a variety of substrates, including other caspase enzymes and neuronal proteins to bind<ref name="ZincMediatedCasp6">PMID: 22891250 </ref>. Furthermore, it is a part of the cysteine-aspartate family[https://en.wikipedia.org/wiki/Caspase], which have these critical amino acid residues in the active site of the enzyme. Caspase-6 has both an inactive zinc-bound conformation and an active ligand-bound conformation, which are largely regulated by variations in zinc concentration<ref name="ZincMediatedCasp6">PMID: 22891250 </ref>. | Found at high concentrations in the brain and bordering tissues, Caspase-6 has been implicated in several neurological diseases including Alzheimer's and dementia[http://www.alz.org/]<ref name="ActiveRegofCasp6andNDdisease">PMID: 25340928 </ref>. It's primarily involved in apoptosis through a largely ambiguous mechanism. It is classified as an endopeptidase[https://en.wikipedia.org/wiki/Endopeptidase] as it cleaves an internal peptide bond of its substrate. It has relatively low specificity in the binding site which allows for a variety of substrates, including other caspase enzymes and neuronal proteins to bind<ref name="ZincMediatedCasp6">PMID: 22891250 </ref>. Furthermore, it is a part of the cysteine-aspartate family[https://en.wikipedia.org/wiki/Caspase], which have these critical amino acid residues in the active site of the enzyme. Caspase-6 has both an inactive zinc-bound conformation and an active ligand-bound conformation, which are largely regulated by variations in zinc concentration<ref name="ZincMediatedCasp6">PMID: 22891250 </ref>. | ||
| + | |||
| + | Caspase-6 is an endopeptidase [https://en.wikipedia.org/wiki/Endopeptidase] involved in apoptosis. In terms of its catalytic function, it is a part of the cysteine-aspartate family [https://en.wikipedia.org/wiki/Caspase]. Before Caspase-6 becomes functional and active, the enzyme exists as a procaspase, also known as a zymogen [https://en.wikipedia.org/wiki/Zymogen][6]. In solution, two zymogens are associated together, forming a homodimer. Zymogen activation, the process by which Caspase-6 becomes active, is largely conserved across the caspase family. | ||
| + | However, Caspase-6 is unique in that it becomes active through self-cleavage rather than cleavage by a separate enzyme. Each zymogen of the unprocessed enzyme contains a <scene name='75/752344/Caspase-6_small_subunit/1'>small</scene> consisting of two helices and <scene name='75/752344/Caspase-6_large_real/1'>large</scene> subunit consisting of three helices, a <scene name='75/752344/Caspase-6_pro-domain/1'>pro-domain</scene>, as well as an intersubunit linker. The helices surround a beta sheet core. In order to become active, the intersubunit linker is bound to the active site of Caspase-6, where it is then cleaved. After cleavage, the four processed subunits, two originating from each zymogen, remain closely associated together through intermolecular forces, forming a dimer of dimers. | ||
[[Image:Caspase-6 protein.jpg|100 px|left|thumb|Caspase-6 Protein]] | [[Image:Caspase-6 protein.jpg|100 px|left|thumb|Caspase-6 Protein]] | ||
Revision as of 23:31, 17 April 2017
Caspase-6 in Homo sapiens
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Wang XJ, Cao Q, Zhang Y, Su XD. Activation and regulation of caspase-6 and its role in neurodegenerative diseases. Annu Rev Pharmacol Toxicol. 2015;55:553-72. doi:, 10.1146/annurev-pharmtox-010814-124414. Epub 2014 Oct 17. PMID:25340928 doi:http://dx.doi.org/10.1146/annurev-pharmtox-010814-124414
- ↑ 2.0 2.1 Velazquez-Delgado EM, Hardy JA. Zinc-Mediated Allosteric Inhibition of Caspase-6. J Biol Chem. 2012 Aug 13. PMID:22891250 doi:http://dx.doi.org/10.1074/jbc.M112.397752
- ↑ Wang XJ, Cao Q, Liu X, Wang KT, Mi W, Zhang Y, Li LF, Leblanc AC, Su XD. Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation. EMBO Rep. 2010 Oct 1. PMID:20890311 doi:10.1038/embor.2010.141
- ↑ Velazquez-Delgado EM, Hardy JA. Phosphorylation regulates assembly of the caspase-6 substrate-binding groove. Structure. 2012 Apr 4;20(4):742-51. Epub 2012 Apr 3. PMID:22483120 doi:10.1016/j.str.2012.02.003