User:Luke Edward Severinac/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
<StructureSection load='4FXO' size='340' side='right' caption='Caspase-6' scene=''> | <StructureSection load='4FXO' size='340' side='right' caption='Caspase-6' scene=''> | ||
| - | Caspase-6 is an [https://en.wikipedia.org/wiki/Endopeptidase endopeptidase] involved in apoptosis. In terms of its catalytic function, it is a part of the [https://en.wikipedia.org/wiki/Caspase cysteine-aspartate family]. Before Caspase-6 becomes functional and active, the enzyme exists as a procaspase, also known as a [https://en.wikipedia.org/wiki/Zymogen zymogen | + | Caspase-6 is an [https://en.wikipedia.org/wiki/Endopeptidase endopeptidase] involved in apoptosis. In terms of its catalytic function, it is a part of the [https://en.wikipedia.org/wiki/Caspase cysteine-aspartate family]. Before Caspase-6 becomes functional and active, the enzyme exists as a procaspase, also known as a [https://en.wikipedia.org/wiki/Zymogen zymogen]. In solution, two zymogens are associated together, forming a homodimer. Zymogen activation, the process by which Caspase-6 becomes active, is largely conserved across the caspase family. |
However, Caspase-6 is unique in that it becomes active through self-cleavage rather than cleavage by a separate enzyme. Each zymogen of the unprocessed enzyme contains a small subunit consisting of two helices and large subunit consisting of three helices, a prodomain, as well as an intersubunit linker. The helices surround a beta sheet core. In order to become active, the intersubunit linker is bound to the active site of Caspase-6, where it is then cleaved. After cleavage, the four processed subunits, two originating from each zymogen, remain closely associated together through intermolecular forces, forming a dimer of dimers. | However, Caspase-6 is unique in that it becomes active through self-cleavage rather than cleavage by a separate enzyme. Each zymogen of the unprocessed enzyme contains a small subunit consisting of two helices and large subunit consisting of three helices, a prodomain, as well as an intersubunit linker. The helices surround a beta sheet core. In order to become active, the intersubunit linker is bound to the active site of Caspase-6, where it is then cleaved. After cleavage, the four processed subunits, two originating from each zymogen, remain closely associated together through intermolecular forces, forming a dimer of dimers. | ||
| Line 8: | Line 8: | ||
=='''Structure'''== | =='''Structure'''== | ||
| + | |||
| + | == Zymogen Activation == | ||
| + | |||
| + | In addition to a self-cleavage mechanism, Caspase-6 zymogen can be activated getting cleaved by Caspase-3, as well as other enzymes. The mechanism of activation by clevage is highly conserved across the caspase family; Self-processing is uniquely recognized as the primary mechanism for Caspase-6 activation, where clevage must occur at two sites for complete activation, specifically the pro-domain and the intersubunit linker. These cleavages are both sequence specific and ordered. First, pro-domain must be cleaved. (Some residues of the pro-domain are not visible in the crystallized structure) Then cleavage of the intersubunit linker occurs, cleaving both DVVD179 and TEVD193. To some extent the pro-domain inhibits Caspase-6's ability to cleave the intersubunit loop and self-activate; It has been proposed that this sequence of cleavage is due to the pro-domain being more readily available to enter the active site. The result of the TETD23 cleavage site priority is that the prodomain acts as a “suicide protector”, which protects the TEVD193 cleavage site from self-cleavage[3]. This protection is necessary when there are low levels of inactive proteins, which must be preserved, in the tissue. The intramolecular cleavage of TETD23 and DVVD179 or TEVD193 are essential for the initiation caspase-6 activation without other caspases present. After both cleavages occur, the processed Caspase-6 can be found in solution as a dimer of dimers. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
===Sequence=== | ===Sequence=== | ||
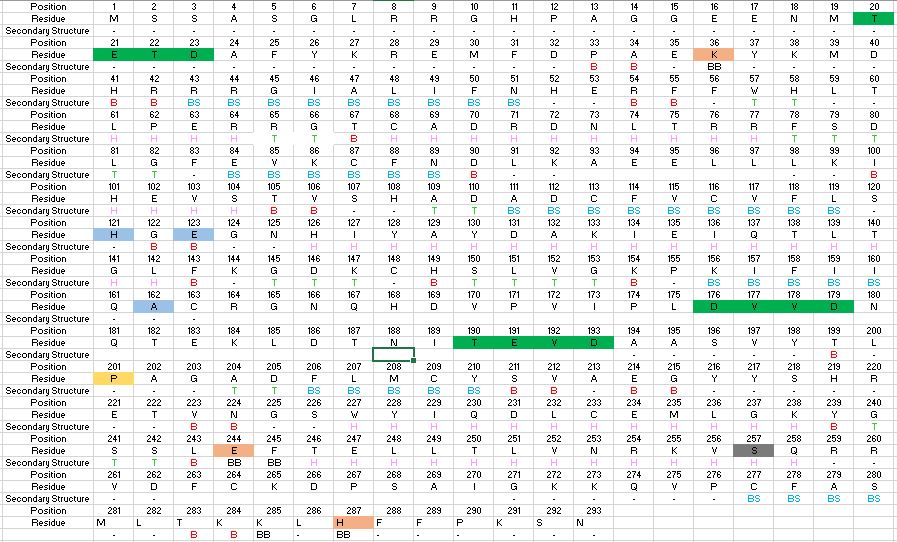
The image below describes the amino acid sequence of Caspase-6, highlighting critical amino acids and sequences necessary for function. It also highlights the secondary structures, which make up the folded protein, and sequences which become cleaved during zymogen processing. | The image below describes the amino acid sequence of Caspase-6, highlighting critical amino acids and sequences necessary for function. It also highlights the secondary structures, which make up the folded protein, and sequences which become cleaved during zymogen processing. | ||
Revision as of 00:15, 18 April 2017
Caspase-6 in Homo sapiens
| |||||||||||
References
- ↑ Wang XJ, Cao Q, Liu X, Wang KT, Mi W, Zhang Y, Li LF, Leblanc AC, Su XD. Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation. EMBO Rep. 2010 Oct 1. PMID:20890311 doi:10.1038/embor.2010.141
- ↑ Velazquez-Delgado EM, Hardy JA. Phosphorylation regulates assembly of the caspase-6 substrate-binding groove. Structure. 2012 Apr 4;20(4):742-51. Epub 2012 Apr 3. PMID:22483120 doi:10.1016/j.str.2012.02.003
- ↑ 3.0 3.1 Wang XJ, Cao Q, Zhang Y, Su XD. Activation and regulation of caspase-6 and its role in neurodegenerative diseases. Annu Rev Pharmacol Toxicol. 2015;55:553-72. doi:, 10.1146/annurev-pharmtox-010814-124414. Epub 2014 Oct 17. PMID:25340928 doi:http://dx.doi.org/10.1146/annurev-pharmtox-010814-124414