User:Natalie Van Ochten/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
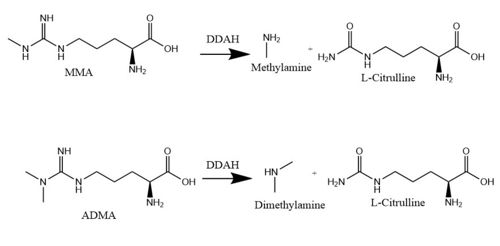
Dimethylarginine Dimethyaminohydrolase <span class="plainlinks">[http://www.chem.qmul.ac.uk/iubmb/enzyme/EC3/5/3/18.html EC 3.5.3.18]</span> (commonly known as DDAH) is a member of the <span class="plainlinks">[https://en.wikipedia.org/wiki/Hydrolase hydrolase]</span> family of enzymes which use water to break down molecules <ref name="palm">Palm F, Onozato ML, Luo Z, Wilcox CS. Dimethylarginine dimethylaminohydrolase (DDAH): expression, regulation, and function in the cardiovascular and renal systems. American Journal of Physiology. 2007 Dec 1;293(6):3227-3245. PMID:<span class="plainlinks">[https://www.ncbi.nlm.nih.gov/pubmed/17933965 17933965]</span> doi:<span class="plainlinks">[http://ajpheart.physiology.org/content/293/6/H3227 10.1152/ajpheart.00998.2007]</span></ref>. Additionally, DDAH is a <span class="plainlinks">[https://en.wikipedia.org/wiki/Nitric_oxide_synthase nitric oxide synthase (NOS)]</span> regulator. It metabolizes free arginine derivatives, namely <span class="plainlinks">[https://en.wikipedia.org/wiki/Asymmetric_dimethylarginine N<sup>Ѡ</sup>,N<sup>Ѡ</sup>-dimethyl-L-arginine (ADMA)]</span> and <span class="plainlinks">[https://en.wikipedia.org/wiki/Methylarginine N<sup>Ѡ</sup>-methyl-L-arginine (MMA)]</span> which competitively inhibit NOS <ref name="tran">Tran CTL, Leiper JM, Vallance P. The DDAH/ADMA/NOS pathway. Atherosclerosis Supplements. 2003 Dec;4(4):33-40. PMID:<span class="plainlinks">[https://www.ncbi.nlm.nih.gov/pubmed/14664901 14664901]</span> doi:<span class="plainlinks">[http://www.sciencedirect.com/science/article/pii/S1567568803000321 10.1016/S1567-5688(03)00032-1]</span></ref>. DDAH converts MMA or ADMA to two products: <span class="plainlinks">[https://en.wikipedia.org/wiki/Citrulline L-citrulline]</span> and an amine <ref name="frey">Frey D, Braun O, Briand C, Vasak M, Grutter MG. Structure of the mammalian NOS regulator dimethylarginine dimethylaminohydrolase: a basis for the design of specific inhibitors. Structure. 2006 May;14(5):901-911. PMID:<span class="plainlinks">[https://www.ncbi.nlm.nih.gov/pubmed/16698551 16698551]</span> doi:<span class="plainlinks">[http://www.sciencedirect.com/science/article/pii/S0969212606001717 10.1016/j.str.2006.03.006]</span></ref> (Figure 1). DDAH is expressed in the cytosol of cells in humans, mice, rates, sheep, cattle, and bacteria <ref name="palm" />. DDAH activity has been localized mainly to the brain, kidney, pancreas, and liver in these organisms. Presented in this page is information from DDAH isoform 1; however, there are two different isoforms <ref name="frey" />. | Dimethylarginine Dimethyaminohydrolase <span class="plainlinks">[http://www.chem.qmul.ac.uk/iubmb/enzyme/EC3/5/3/18.html EC 3.5.3.18]</span> (commonly known as DDAH) is a member of the <span class="plainlinks">[https://en.wikipedia.org/wiki/Hydrolase hydrolase]</span> family of enzymes which use water to break down molecules <ref name="palm">Palm F, Onozato ML, Luo Z, Wilcox CS. Dimethylarginine dimethylaminohydrolase (DDAH): expression, regulation, and function in the cardiovascular and renal systems. American Journal of Physiology. 2007 Dec 1;293(6):3227-3245. PMID:<span class="plainlinks">[https://www.ncbi.nlm.nih.gov/pubmed/17933965 17933965]</span> doi:<span class="plainlinks">[http://ajpheart.physiology.org/content/293/6/H3227 10.1152/ajpheart.00998.2007]</span></ref>. Additionally, DDAH is a <span class="plainlinks">[https://en.wikipedia.org/wiki/Nitric_oxide_synthase nitric oxide synthase (NOS)]</span> regulator. It metabolizes free arginine derivatives, namely <span class="plainlinks">[https://en.wikipedia.org/wiki/Asymmetric_dimethylarginine N<sup>Ѡ</sup>,N<sup>Ѡ</sup>-dimethyl-L-arginine (ADMA)]</span> and <span class="plainlinks">[https://en.wikipedia.org/wiki/Methylarginine N<sup>Ѡ</sup>-methyl-L-arginine (MMA)]</span> which competitively inhibit NOS <ref name="tran">Tran CTL, Leiper JM, Vallance P. The DDAH/ADMA/NOS pathway. Atherosclerosis Supplements. 2003 Dec;4(4):33-40. PMID:<span class="plainlinks">[https://www.ncbi.nlm.nih.gov/pubmed/14664901 14664901]</span> doi:<span class="plainlinks">[http://www.sciencedirect.com/science/article/pii/S1567568803000321 10.1016/S1567-5688(03)00032-1]</span></ref>. DDAH converts MMA or ADMA to two products: <span class="plainlinks">[https://en.wikipedia.org/wiki/Citrulline L-citrulline]</span> and an amine <ref name="frey">Frey D, Braun O, Briand C, Vasak M, Grutter MG. Structure of the mammalian NOS regulator dimethylarginine dimethylaminohydrolase: a basis for the design of specific inhibitors. Structure. 2006 May;14(5):901-911. PMID:<span class="plainlinks">[https://www.ncbi.nlm.nih.gov/pubmed/16698551 16698551]</span> doi:<span class="plainlinks">[http://www.sciencedirect.com/science/article/pii/S0969212606001717 10.1016/j.str.2006.03.006]</span></ref> (Figure 1). DDAH is expressed in the cytosol of cells in humans, mice, rates, sheep, cattle, and bacteria <ref name="palm" />. DDAH activity has been localized mainly to the brain, kidney, pancreas, and liver in these organisms. Presented in this page is information from DDAH isoform 1; however, there are two different isoforms <ref name="frey" />. | ||
| - | [[Image:DDAH mechanism.jpg|500 px|center|thumb|Figure 1.]] | + | [[Image:DDAH mechanism.jpg|500 px|center|thumb|Figure 1. The normal DDAH mechanism]] |
==Different Isoforms== | ==Different Isoforms== | ||
| Line 13: | Line 13: | ||
===Lid Region=== | ===Lid Region=== | ||
| - | Amino acids 25-36 of DDAH constitute the loop region of the protein which is more commonly known as the lid region <ref name="frey" /> | + | Amino acids 25-36 of DDAH constitute the loop region of the protein which is more commonly known as the lid region <ref name="frey" />. The lid is what allows the active site to be exposed to substrate binding or not. Studies have shown crystal structures of the lid at <scene name='69/694225/Low_ph_w_lid/1'>open</scene> and <scene name='69/694225/Closed_lid_zn9/1'>closed </scene> conformations. In the open conformation, the lid forms an alpha helix and the amino acid <scene name='69/694225/Low_ph_w_lid/2'>Leu29</scene> is moved so it does not interact with the active site. This allows the active site to be vulnerable to attack. This lid region is very flexible. This open conformation has been shown when DDAH had been <span class="plainlinks">[https://en.wikipedia.org/wiki/Crystallization crystallized]</span> when <span class="plainlinks">[https://en.wikipedia.org/wiki/Zinc Zn(II)]</span> was bound at <scene name='69/694225/Active_site6/1'>pH 6.3</scene>. There is a closed form which has been observed with Zn(II) binding at <scene name='69/694225/Active_site_9/1'>pH 9.0</scene> and in the unliganded enzyme. When the lid is closed, a specific <scene name=’75/752351/Hbond_leu29/1’>hydrogen bond</scene> can form between the Leu29 carbonyl and the amino group on bound molecule. This stabilizes this complex. The Leu29 is then <scene name='69/694225/Closed_lid_zn9/3'>blocking</scene> the active site entrance <ref name="frey" />. Opening and closing the lid takes place faster than the actual reaction in the active site <ref name="rasheed">Rasheed M, Richter C, Chisty LT, Kirkpatrick J, Blackledge M, Webb MR, Driscoll PC. Ligand-dependent dynamics of the active site lid in bacterial Dimethyarginine Dimethylaminohydrolase. Biochemistry. 2014 Feb 18;53:1092-1104. PMCID:<span class="plainlinks">[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3945819/ PMC3945819]</span> doi:<span class="plainlinks">[http://pubs.acs.org/doi/abs/10.1021/bi4015924 10.1021/bi4015924]</span></ref>. This suggests that the <span class="plainlinks">[https://en.wikipedia.org/wiki/Rate-determining_step rate-limiting step]</span> of this reaction is not the lid movement but is the actual chemistry happening to the substrate in the active site of DDAH <ref name="rasheed" />. |
| - | The specific residues in the lid region are different in different organisms <ref name="frey" />. The only consistent similarity is a <span class="plainlinks">[https://en.wikipedia.org/wiki/Conserved_sequence conserved]</span> leucine residue in this lid that function to hydrogen bond with the <span class="plainlinks">[https://en.wikipedia.org/wiki/Ligand ligand]</span> bound to the active site <ref name="rasheed" /> (Figure 2). Different <span class="plainlinks">[https://en.wikipedia.org/wiki/Protein_isoform isoforms]</span> from the same species can have differences in lid regions as well <ref name="frey" />. DDAH-2 has a negatively charged lid while DDAH-1 has a positively charged lid <ref name="frey" />. | + | The specific residues in the lid region are different in different organisms <ref name="frey" /> (Figure 2). The only consistent similarity is a <span class="plainlinks">[https://en.wikipedia.org/wiki/Conserved_sequence conserved]</span> leucine residue in this lid that function to hydrogen bond with the <span class="plainlinks">[https://en.wikipedia.org/wiki/Ligand ligand]</span> bound to the active site in DDAH-1 but not in DDAH-2 <ref name="rasheed" /> (Figure 2). Different <span class="plainlinks">[https://en.wikipedia.org/wiki/Protein_isoform isoforms]</span> from the same species can have differences in lid regions as well <ref name="frey" />. DDAH-2 has a negatively charged lid while DDAH-1 has a positively charged lid <ref name="frey" />. |
| - | [[Image:Lid Region WebLogo.png|500 px|center|thumb|Figure 2.]] | + | [[Image:Lid Region WebLogo.png|500 px|center|thumb|Figure 2. WebLogo for the lid region in DDAH-1 of many different organisms.]] |
===Active Site=== | ===Active Site=== | ||
| - | The normal DDAH regulation <span class="plainlinks">[https://en.wikipedia.org/wiki/Reaction_mechanism mechanism]</span> depends on the presence of <scene name='75/752351/Ddah_active_site/2'>Cys 249</scene> in the active site that acts as a <span class="plainlinks">[https://en.wikipedia.org/wiki/Nucleophile nucleophile]</span> in the mechanism <ref name="stone">Stone EM, Costello AL, Tierney DL, Fast W. Substrate-assisted cysteine deprotonation in the mechanism of Dimethylargininase (DDAH) from Pseudomonas aeruginosa. Biochemistry. 2006 May 2;45(17):5618-5630. PMID:<span class="plainlinks">[https://www.ncbi.nlm.nih.gov/pubmed/16634643 16634643]</span> doi:<span class="plainlinks">[http://pubs.acs.org/doi/abs/10.1021/bi052595m 10.1021/bi052595m]</span></ref>. The Cys249 is used to attack the <span class="plainlinks">[https://en.wikipedia.org/wiki/Guanidine guanidinium]</span> carbon on the substrate that is held in the active site via hydrogen bonds | + | The normal DDAH regulation <span class="plainlinks">[https://en.wikipedia.org/wiki/Reaction_mechanism mechanism]</span> depends on the presence of <scene name='75/752351/Ddah_active_site/2'>Cys 249</scene> in the active site that acts as a <span class="plainlinks">[https://en.wikipedia.org/wiki/Nucleophile nucleophile]</span> in the mechanism <ref name="stone">Stone EM, Costello AL, Tierney DL, Fast W. Substrate-assisted cysteine deprotonation in the mechanism of Dimethylargininase (DDAH) from Pseudomonas aeruginosa. Biochemistry. 2006 May 2;45(17):5618-5630. PMID:<span class="plainlinks">[https://www.ncbi.nlm.nih.gov/pubmed/16634643 16634643]</span> doi:<span class="plainlinks">[http://pubs.acs.org/doi/abs/10.1021/bi052595m 10.1021/bi052595m]</span></ref> (Figure 3). The Cys249 is used to attack the <span class="plainlinks">[https://en.wikipedia.org/wiki/Guanidine guanidinium]</span> carbon on the substrate that is held in the active site via hydrogen bonds. This is followed by collapsing the tetrahedral product to get rid of the <span class="plainlinks">[https://en.wikipedia.org/wiki/Alkylamines alkylamine]</span> leaving group. A <span class="plainlinks">[https://en.wikipedia.org/wiki/Isothiouronium thiouronium]</span> intermediate is then formed with <span class="plainlinks">[https://en.wikipedia.org/wiki/Orbital_hybridisation sp<sup>2</sup> hybridization]</span>. This intermediate is hydrolyzed to form citrulline. The <scene name='75/752351/Ddah_active_site_his162/1'>His162</scene> protonates the leaving group in this reaction and generates hydroxide to hydrolyze the intermediate formed in the reaction (Figure 3). Studies suggest that Cys249 is neutral until binding of guanidinium near Cys249 decreases Cys249’s <span class="plainlinks">[https://en.wikipedia.org/wiki/Acid_dissociation_constant pKa]</span> and deprotonates the thiolate to activate the nucleophile. Other studies suggest that the Cys249 and an active site His162 form an <span class="plainlinks">[https://en.wikipedia.org/wiki/Intimate_ion_pair ion pair]</span> to deprotonate the thiolate. Cys249 and His162 can also form a binding site for inhibitors to bind to which stabilizes the thiolate. This is important in regulating NO activity in organisms and designing drugs to inhibit this enzyme <ref name="stone" />. |
| - | [[Image:The Normal DDAH Mechanism.jpg|800px|center|thumb|Figure 3.]] | + | [[Image:The Normal DDAH Mechanism.jpg|800px|center|thumb|Figure 3. The normal mechanism of DDAH highlighting important residues involved.]] |
| - | There is a channel in the center of the protein that is closed by a <scene name='75/752351/Ddah_salt_bridge/3'>salt bridge</scene> connecting Glu77 and Lys174 <ref name="frey" />. This salt bridge constitutes the bottom of the active site. There is a pore containing water on one side of the channel. This pore is delineated by the first β strand of each of the five propeller blades. The water in the water-filled pore forms hydrogen bonds to His172 and Ser175. The other side of the channel is the active site. Short loop regions and a helical structure define the outward boundaries of this site. Active sites of DDAH from different organisms | + | There is a channel in the center of the protein that is closed by a <scene name='75/752351/Ddah_salt_bridge/3'>salt bridge</scene> connecting Glu77 and Lys174 <ref name="frey" />. This salt bridge constitutes the bottom of the active site. There is a pore containing water on one side of the channel. This pore is delineated by the first β strand of each of the five propeller blades. The water in the water-filled pore forms hydrogen bonds to His172 and Ser175. The other side of the channel is the active site. Short loop regions and a helical structure define the outward boundaries of this site. Active sites of DDAH from different organisms are similar. Amino acids involved in the chemical mechanism of creating products are also <scene name='69/694225/Evolutionary_conservation/1'>conserved</scene> (Figure 4). |
| - | Amino acids in the lid region are not conserved except for a Leucine amino acid. When MMA or ADMA bind in the active site, they are broken down into L-citrulline and amines (Figure 1). L-citrulline leaves the active site when the lid opens. The amines can either leave through the entrance to the active site or through a pore made by movement of Glu77 and Lys174 <ref name="frey" />. | + | [[Image:ColorKey ConSurf NoYellow NoGray.gif|400px|right|thumb|Figure 4. Color key for DDAH conservation]] |
| + | Amino acids in the lid region are not conserved except for a Leucine amino acid (Figure 3). When MMA or ADMA bind in the active site, they are broken down into L-citrulline and amines (Figure 1). L-citrulline leaves the active site when the lid opens. The amines can either leave through the entrance to the active site or through a pore made by movement of Glu77 and Lys174 <ref name="frey" />. | ||
====Zn(II) Bound to the Active Site==== | ====Zn(II) Bound to the Active Site==== | ||
| - | In DDAH, Zinc (ZnII) acts as an endogenous inhibitor and prevents normal NOS activity <ref name="frey" />. The Zn(II)-binding site is located inside the protein’s active site, which makes it a <span class="plainlinks">[https://en.wikipedia.org/wiki/Competitive_inhibition competitive inhibitor]</span> | + | In DDAH, Zinc (ZnII) acts as an endogenous inhibitor and prevents normal NOS activity <ref name="frey" /> (Figure 5). The Zn(II)-binding site is located inside the protein’s active site, which makes it a <span class="plainlinks">[https://en.wikipedia.org/wiki/Competitive_inhibition competitive inhibitor]</span>. When bound, Zn(II) blocks the entrance of any other substrate (Figure 5). It was found that Cys273, His172, Glu77, Asp78, and Asp 268 all play a role in the binding of Zn(II). <scene name='69/694225/Active_site6/1'>Cys273</scene> directly coordinates with the Zn(II) ion in the active site while the other significant residues stabilize the ion via hydrogen bonding interactions with water molecules in the active site. Depending on pH, His172 can <scene name='69/694225/Active_site_9/1'>change conformation</scene> and use the <span class="plainlinks">[https://en.wikipedia.org/wiki/Imidazole imidazole]</span> group to directly coordinate the Zn(II) ion. Cys273, which is conserved between bovine and humans, is the key active site residue that coordinates Zn(II) <ref name="frey" />. Zinc-cysteine complexes have been found to be important mediators of protein <span class="plainlinks">[https://en.wikipedia.org/wiki/Catalysis catalysis]</span>, regulation, and structure <ref name="pace">Pace NJ, Weerpana E. Zinc-binding cysteines: diverse functions and structural motifs. Biomolecules. 2014 June;4(2):419-434. PMCID:<span class="plainlinks">[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4101490/ 4101490]</span> doi:<span class="plainlinks">[http://www.mdpi.com/2218-273X/4/2/419/htm 10.3390/biom4020419]</span> </ref>. Cys273 and the water molecules stabilize the Zn(II) ion in a tetrahedral environment. The Zn(II) dissociation constant is 4.2 nM which is consistent with the nanomolar concentrations of Zn(II) in the cells, which provides more evidence for the regulatory use of Zn(II) by DDAH <ref name="pace" />. |
| - | + | [[Image:Zn(II) bound at differing pH values.jpg|500 px|center|thumb|Figure 5. Zn(II) bound to the active site of DDAH at differing pH values. A) Zn(II) bound at pH 9.0 showing the channel of DDAH. B) Zn(II) bound at 9.0 showing the closed conformation lid with Leu29 blocking the active site. C) Zn(II) bound at pH 6.3 showing the channel of DDAH. D) Zn(II) bound at pH 6.3 showing the open lid conformation with Leu29 away from the active site.]] | |
| - | [[Image:Zn(II) bound at differing pH values.jpg| | + | |
===Inhibitors=== | ===Inhibitors=== | ||
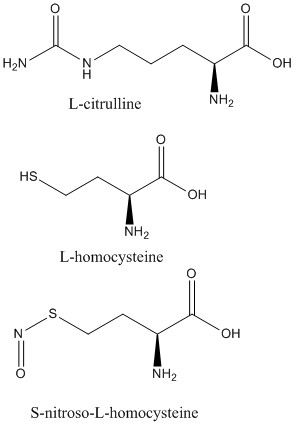
| - | <scene name='75/752351/Ddah_l-homocysteine/2'>L-homocysteine</scene> and <scene name='75/752351/Ddah_with_L-citrulline/4'>L-citrulline</scene> bind in the active site in the same orientation to create the same <span class="plainlinks">[https://en.wikipedia.org/wiki/Intermolecular_force intermolecular bonds]</span> between them and DDAH <ref name="frey" />. L-citrulline is a product of DDAH hydrolyzing ADMA and MMA, suggesting DDAH activity creates a <span class="plainlinks">[https://en.wikipedia.org/wiki/Negative_feedback negative feedback]</span> loop on itself. Both molecules enter the active site and cause DDAH to be in its closed lid formation. The α carbon on either molecule creates three salt bridges with DDAH: two with the guanidine group of Arg144 and one with the guanidine group Arg97. Another salt bridge is formed between the ligand and Asp72. The molecules are stabilized in the active site by hydrogen bonds: α carbon-amino group of the ligand to main chain carbonyls of Val267 and Leu29. Hydrogen bonds also form between the side chains of Asp78 and Glu77 with the ureido group of L-citrulline. | + | <scene name='75/752351/Ddah_l-homocysteine/2'>L-homocysteine</scene> and <scene name='75/752351/Ddah_with_L-citrulline/4'>L-citrulline</scene> bind in the active site in the same orientation to create the same <span class="plainlinks">[https://en.wikipedia.org/wiki/Intermolecular_force intermolecular bonds]</span> between them and DDAH <ref name="frey" /> (Figure 6). L-citrulline is a product of DDAH hydrolyzing ADMA and MMA, suggesting DDAH activity creates a <span class="plainlinks">[https://en.wikipedia.org/wiki/Negative_feedback negative feedback]</span> loop on itself. Both molecules enter the active site and cause DDAH to be in its closed lid formation. The α carbon on either molecule creates three salt bridges with DDAH: two with the guanidine group of Arg144 and one with the guanidine group Arg97. Another salt bridge is formed between the ligand and Asp72. The molecules are stabilized in the active site by hydrogen bonds: α carbon-amino group of the ligand to main chain carbonyls of Val267 and Leu29. Hydrogen bonds also form between the side chains of Asp78 and Glu77 with the ureido group of L-citrulline. |
| - | Like L-homocysteine and L-citrulline, <scene name='75/752351/Ddah_s-nitroso-l-homocysteine/3'>S-nitroso-L-homocysteine</scene> binds and the lid region of DDAH is closed. When DDAH reacts with S-nitroso-L-homocysteine, a covalent product, N-thiosulfximide exist in the active site because of its binding to Cys273. N-thiosulfximide is stabilized by several salt bridges and hydrogen bonds. Arg144 and Arg97 stabilize the α carbon-carbonyl group via salt bridges, and Leu29, Val267, and Asp72 stabilize the α carbon-amino group by forming hydrogen bonds <ref name="frey" />. | + | Like L-homocysteine and L-citrulline, <scene name='75/752351/Ddah_s-nitroso-l-homocysteine/3'>S-nitroso-L-homocysteine</scene> binds and the lid region of DDAH is closed (Figure 6). When DDAH reacts with S-nitroso-L-homocysteine, a covalent product, N-thiosulfximide exist in the active site because of its binding to Cys273. N-thiosulfximide is stabilized by several salt bridges and hydrogen bonds. Arg144 and Arg97 stabilize the α carbon-carbonyl group via salt bridges, and Leu29, Val267, and Asp72 stabilize the α carbon-amino group by forming hydrogen bonds <ref name="frey" />. |
| - | [[Image:L-citrulline, L-homocysteine, and S-nitroso-L-homocysteine.jpg|500px|center|thumb|Figure | + | [[Image:L-citrulline, L-homocysteine, and S-nitroso-L-homocysteine.jpg|500px|center|thumb|Figure 6. Structures of DDAH inhibitors.]] |
==Clinical Relevance== | ==Clinical Relevance== | ||
| Line 51: | Line 51: | ||
== 3D Structures of Dimethylarginine Dimethylaminohydrolase == | == 3D Structures of Dimethylarginine Dimethylaminohydrolase == | ||
| - | <span class="plainlinks">[http://proteopedia.org/wiki/index.php/2c6z 2C6Z]</span> | + | <span class="plainlinks">[http://proteopedia.org/wiki/index.php/2c6z 2C6Z]</span> L-citrulline bound to isoform 1 |
| - | <span class="plainlinks">[http://proteopedia.org/wiki/index.php/2ci1 2CI1]</span> | + | <span class="plainlinks">[http://proteopedia.org/wiki/index.php/2ci1 2CI1]</span> S-nitroso-L-homocysteine bound to isoform 1 |
| - | <span class="plainlinks">[http://proteopedia.org/wiki/index.php/2ci3 2CI3]</span> | + | <span class="plainlinks">[http://proteopedia.org/wiki/index.php/2ci3 2CI3]</span> crystal form 1 |
| - | <span class="plainlinks">[http://proteopedia.org/wiki/index.php/2ci4 2CI4]</span> | + | <span class="plainlinks">[http://proteopedia.org/wiki/index.php/2ci4 2CI4]</span> crystal form 2 |
| - | <span class="plainlinks">[http://proteopedia.org/wiki/index.php/2ci5 2CI5]</span> | + | <span class="plainlinks">[http://proteopedia.org/wiki/index.php/2ci5 2CI5]</span> L-homocysteine bound to isoform 1 |
| - | <span class="plainlinks">[http://proteopedia.org/wiki/index.php/2ci6 2CI6]</span> | + | <span class="plainlinks">[http://proteopedia.org/wiki/index.php/2ci6 2CI6]</span> Zn (II) bound at low pH to isoform 1 |
| - | <span class="plainlinks">[http://proteopedia.org/wiki/index.php/2ci7 2CI7]</span> | + | <span class="plainlinks">[http://proteopedia.org/wiki/index.php/2ci7 2CI7]</span> Zn (II) bound at high pH to isoform 1 |
Revision as of 13:44, 18 April 2017
Dimethylarginine Dimethylaminohydrolase
| |||||||||||
References
- ↑ 1.0 1.1 Palm F, Onozato ML, Luo Z, Wilcox CS. Dimethylarginine dimethylaminohydrolase (DDAH): expression, regulation, and function in the cardiovascular and renal systems. American Journal of Physiology. 2007 Dec 1;293(6):3227-3245. PMID:17933965 doi:10.1152/ajpheart.00998.2007
- ↑ 2.0 2.1 2.2 Tran CTL, Leiper JM, Vallance P. The DDAH/ADMA/NOS pathway. Atherosclerosis Supplements. 2003 Dec;4(4):33-40. PMID:14664901 doi:10.1016/S1567-5688(03)00032-1
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 3.11 3.12 3.13 3.14 3.15 3.16 3.17 3.18 3.19 3.20 3.21 3.22 3.23 Frey D, Braun O, Briand C, Vasak M, Grutter MG. Structure of the mammalian NOS regulator dimethylarginine dimethylaminohydrolase: a basis for the design of specific inhibitors. Structure. 2006 May;14(5):901-911. PMID:16698551 doi:10.1016/j.str.2006.03.006
- ↑ Humm A, Fritsche E, Mann K, Göhl M, Huber R. Recombinant expression and isolation of human L-arginine:glycine amidinotransferase and identification of its active-site cysteine residue. Biochemical Journal. 1997 March 15;322(3):771-776. PMID:9148748 doi:10.1042/bj3220771
- ↑ 5.0 5.1 5.2 Rasheed M, Richter C, Chisty LT, Kirkpatrick J, Blackledge M, Webb MR, Driscoll PC. Ligand-dependent dynamics of the active site lid in bacterial Dimethyarginine Dimethylaminohydrolase. Biochemistry. 2014 Feb 18;53:1092-1104. PMCID:PMC3945819 doi:10.1021/bi4015924
- ↑ 6.0 6.1 Stone EM, Costello AL, Tierney DL, Fast W. Substrate-assisted cysteine deprotonation in the mechanism of Dimethylargininase (DDAH) from Pseudomonas aeruginosa. Biochemistry. 2006 May 2;45(17):5618-5630. PMID:16634643 doi:10.1021/bi052595m
- ↑ 7.0 7.1 Pace NJ, Weerpana E. Zinc-binding cysteines: diverse functions and structural motifs. Biomolecules. 2014 June;4(2):419-434. PMCID:4101490 doi:10.3390/biom4020419
- ↑ Janssen W, Pullamsetti SS, Cooke J, Weissmann N, Guenther A, Schermuly RT. The role of dimethylarginine dimethylaminohydrolase (DDAH) in pulmonary fibrosis. The Journal of Pathology. 2012 Dec 12;229(2):242-249. Epub 2013 Jan. PMID:23097221 doi:10.1002/path.4127
- ↑ Colasanti M, Suzuki H. The dual personality of NO. ScienceDirect. 2000 Jul 1;21(7):249-252. PMID:10979862 doi:10.1016/S0165-6147(00)01499-1
- ↑ Rassaf T, Feelisch M, Kelm M. Circulating NO pool: assessment of nitrite and nitroso species in blood and tissues. Free Rad. Biol. Med. 2004 Feb 15;36(4):413-422. PMID:14975444 doi:10.1016/j.freeradbiomed.2003.11.011
- ↑ Tsao PS, Cooke JP. Endothelial alterations in hypercholesterolemia: more than simply vasodilator dysfunction. Journal of Cardiovascular Pharmacology. 1998;32(3):48-53. PMID:9883748
- ↑ Vallance P, Leiper J. Blocking NO synthesis: how, where and why? Nat. Rev. Drug Discov. 2002 Dec;1(12):939-950. PMID:12461516 doi:10.1038/nrd960
Student Contributors
- Natalie Van Ochten
- Kaitlyn Enderle
- Colton Junod
3D Structures of Dimethylarginine Dimethylaminohydrolase
2C6Z L-citrulline bound to isoform 1
2CI1 S-nitroso-L-homocysteine bound to isoform 1
2CI3 crystal form 1
2CI4 crystal form 2
2CI5 L-homocysteine bound to isoform 1
2CI6 Zn (II) bound at low pH to isoform 1
2CI7 Zn (II) bound at high pH to isoform 1