We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1063
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
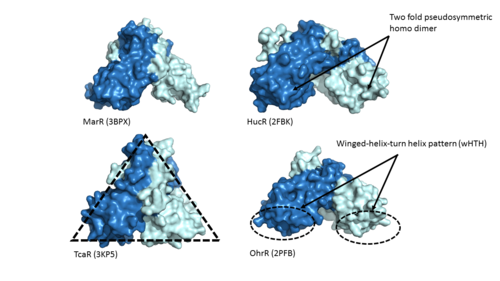
Adhesin Competence Regulator (AdcR) is a transcriptional regulator that controls the activation of over seventy genes within the bacteria [https://en.wikipedia.org/wiki/Streptococcus_pneumoniae''Streptococcus pneumoniae''] and is a member of the multiple antibiotic resistance regulator (MarR) protein family <ref>DOI:10.1093/nar/gku1304 </ref>. Members of the MarR protein family conserve a number of features including a general triangular shape, a two fold pseudosymmetric homodimer, and a winged helix-turn-helix pattern [https://en.wikipedia.org/wiki/Helix-turn-helix (wHTH)] which can be seen in Figure 1. Consistent with AdcR's identity as a member of the MarR protein family, AdcR exhibits these conserved features. Additionally this structure calls for multiple zinc binding sites that facilitate protein conformational change allowing for DNA binding and regulation through the wHTH domain. | Adhesin Competence Regulator (AdcR) is a transcriptional regulator that controls the activation of over seventy genes within the bacteria [https://en.wikipedia.org/wiki/Streptococcus_pneumoniae''Streptococcus pneumoniae''] and is a member of the multiple antibiotic resistance regulator (MarR) protein family <ref>DOI:10.1093/nar/gku1304 </ref>. Members of the MarR protein family conserve a number of features including a general triangular shape, a two fold pseudosymmetric homodimer, and a winged helix-turn-helix pattern [https://en.wikipedia.org/wiki/Helix-turn-helix (wHTH)] which can be seen in Figure 1. Consistent with AdcR's identity as a member of the MarR protein family, AdcR exhibits these conserved features. Additionally this structure calls for multiple zinc binding sites that facilitate protein conformational change allowing for DNA binding and regulation through the wHTH domain. | ||
| - | [[Image:MarR_protein_family_slide.png|500px|left|thumb|'''Figure 1'''Proteins MarR [http://www.rcsb.org/pdb/explore/explore.do?structureId=3bpx (3BPX)], HucR [http://www.rcsb.org/pdb/explore/explore.do?structureId=2FBK (2FBK)], TcaR [http://www.rcsb.org/pdb/explore/explore.do?structureId=3KP5 (3KP5)], and OhrR [http://www.rcsb.org/pdb/explore/explore.do?structureId=2pfb (2PFB)] are pictured above with conserved features of the MarR protein family highlighted]] | + | [[Image:MarR_protein_family_slide.png|500px|left|thumb|'''Figure 1'''. Proteins MarR [http://www.rcsb.org/pdb/explore/explore.do?structureId=3bpx (3BPX)], HucR [http://www.rcsb.org/pdb/explore/explore.do?structureId=2FBK (2FBK)], TcaR [http://www.rcsb.org/pdb/explore/explore.do?structureId=3KP5 (3KP5)], and OhrR [http://www.rcsb.org/pdb/explore/explore.do?structureId=2pfb (2PFB)] are pictured above with conserved features of the MarR protein family highlighted]] |
Zinc plays a vital role in organism homeostasis, acting as a [https://en.wikipedia.org/wiki/Cofactor_(biochemistry) co-factor] and a regulator of enzymatic activity. However zinc can lead to cell toxicity and deficiency of other vital metals that are also necessary for protein function <ref> Fraústo da Silva J, Williams R. The Biological Chemistry of Elements: The Inorganic Chemistry of Life. Second ed. Oxford University Press; Oxford: 2001.</ref><ref> DOI: 10.1021/cr900077w</ref>. The importance of AdcR in ''Streptococcus pneumoniae'' can be understood provided its ability to regulate zinc transfer proteins within the bacteria. Contrasting with other members of the MarR family, AdcR is metal dependent. Binding of Zinc allows AdcR to bind DNA and activate the transcription of high-affinity Zinc specific uptake transporters. The binding of Zinc induces a conformational change that allows for a hydrogen bond network between helices of the binding domain. It is believed that this hydrogen bond network is the allosteric activator needed to expose residues that bind the bases along the major groove of the DNA <ref name="guerra">PMID:22085181</ref>. | Zinc plays a vital role in organism homeostasis, acting as a [https://en.wikipedia.org/wiki/Cofactor_(biochemistry) co-factor] and a regulator of enzymatic activity. However zinc can lead to cell toxicity and deficiency of other vital metals that are also necessary for protein function <ref> Fraústo da Silva J, Williams R. The Biological Chemistry of Elements: The Inorganic Chemistry of Life. Second ed. Oxford University Press; Oxford: 2001.</ref><ref> DOI: 10.1021/cr900077w</ref>. The importance of AdcR in ''Streptococcus pneumoniae'' can be understood provided its ability to regulate zinc transfer proteins within the bacteria. Contrasting with other members of the MarR family, AdcR is metal dependent. Binding of Zinc allows AdcR to bind DNA and activate the transcription of high-affinity Zinc specific uptake transporters. The binding of Zinc induces a conformational change that allows for a hydrogen bond network between helices of the binding domain. It is believed that this hydrogen bond network is the allosteric activator needed to expose residues that bind the bases along the major groove of the DNA <ref name="guerra">PMID:22085181</ref>. | ||
| Line 14: | Line 14: | ||
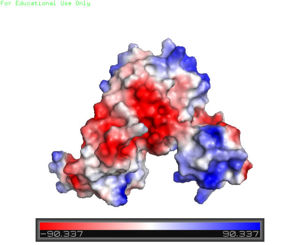
The <scene name='69/694230/Whth_2/1'>winged helix-turn-helix</scene> motif is made up of the <font color='blue'>alpha 2</font> and <font color='blue'>alpha 4 helices</font> along with <scene name='69/694230/Anti-parallel_beta_sheet/1'>anti-parallel beta sheets</scene> on each side. Only one monomer is shown for clarity purposes. There is one wHTH motif per monomer. The recognition helix, or the alpha 4 helix, binds the major groove of DNA through hydrogen bonding and Van der Waals interactions between exposed bases. The wings of the helix bind the minor groove of DNA while the other helices stabilize the DNA and Protein upon binding. The two anti parallel beta sheets contain several <scene name='69/694230/Positive_residues_on_wing/5'>Arginine, Asparagine, and Lysine residues</scene> that stabilize this interaction between DNA. The charge map down below highlights the dark blue tips consisting of lysine and arginine residues, which stabilize the negatively charged backbone of DNA. | The <scene name='69/694230/Whth_2/1'>winged helix-turn-helix</scene> motif is made up of the <font color='blue'>alpha 2</font> and <font color='blue'>alpha 4 helices</font> along with <scene name='69/694230/Anti-parallel_beta_sheet/1'>anti-parallel beta sheets</scene> on each side. Only one monomer is shown for clarity purposes. There is one wHTH motif per monomer. The recognition helix, or the alpha 4 helix, binds the major groove of DNA through hydrogen bonding and Van der Waals interactions between exposed bases. The wings of the helix bind the minor groove of DNA while the other helices stabilize the DNA and Protein upon binding. The two anti parallel beta sheets contain several <scene name='69/694230/Positive_residues_on_wing/5'>Arginine, Asparagine, and Lysine residues</scene> that stabilize this interaction between DNA. The charge map down below highlights the dark blue tips consisting of lysine and arginine residues, which stabilize the negatively charged backbone of DNA. | ||
| - | [[Image:Charge_map.jpg |300 px|right|thumb|'''Figure 2''' A charge map of AdcR shows the general triangular shape and the positive charged (blue) area on HTH domains]] | + | [[Image:Charge_map.jpg |300 px|right|thumb|'''Figure 2'''. A charge map of AdcR shows the general triangular shape and the positive charged (blue) area on HTH domains]] |
=== Hydrogen Bond Network === | === Hydrogen Bond Network === | ||
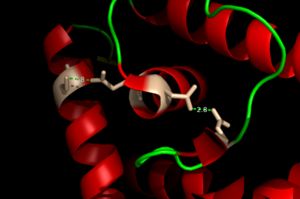
The binding of Zinc allows for the conformational change that induces the binding of DNA in order to activate genes. The binding of Zinc metals creates a hydrogen bond network within the protein that connects the metal binding sites and the [https://en.wikipedia.org/wiki/DNA-binding_domain DNA binding domain]. More importantly, the hydrogen bonding network connects the metal binding pockets to the alpha 4 helix. Alpha 4 helix on each monomer plays a crucial role in binding DNA because it acts as the recognition helix. <scene name='69/694230/Recognition_helix/2'>Specific residues</scene> in the recognition helix recognize a sequence of DNA that is unknown at the moment; however, scientists do know that the hydrogen bond network acts as an allosteric activator for the protein to bind DNA. The hydrogen bond network connects the alpha 2 and alpha 4 helix via hydrogen bonding between specific residues. After zinc is bound, a glutamate (E24) residue from a random coil accepts a hydrogen bond from the carboxamide end of an asparagine (N38) residue from the alpha 2 helix. Then, a glutamine (Q40) residue from alpha 2 helix accepts a hydrogen bond from a serine (S74) residue from the alpha 4 helix <ref name="guerra" />. The <scene name='69/694230/Hydrogen_bonding_1/3'>hydrogen bond network</scene> (<scene name='69/694230/Hydrogen_bonding_2/2'>with measurements</scene>) is represented by each atom type in the 3D model. The hydrogen bond network is characteristic of the MarR family as a whole. | The binding of Zinc allows for the conformational change that induces the binding of DNA in order to activate genes. The binding of Zinc metals creates a hydrogen bond network within the protein that connects the metal binding sites and the [https://en.wikipedia.org/wiki/DNA-binding_domain DNA binding domain]. More importantly, the hydrogen bonding network connects the metal binding pockets to the alpha 4 helix. Alpha 4 helix on each monomer plays a crucial role in binding DNA because it acts as the recognition helix. <scene name='69/694230/Recognition_helix/2'>Specific residues</scene> in the recognition helix recognize a sequence of DNA that is unknown at the moment; however, scientists do know that the hydrogen bond network acts as an allosteric activator for the protein to bind DNA. The hydrogen bond network connects the alpha 2 and alpha 4 helix via hydrogen bonding between specific residues. After zinc is bound, a glutamate (E24) residue from a random coil accepts a hydrogen bond from the carboxamide end of an asparagine (N38) residue from the alpha 2 helix. Then, a glutamine (Q40) residue from alpha 2 helix accepts a hydrogen bond from a serine (S74) residue from the alpha 4 helix <ref name="guerra" />. The <scene name='69/694230/Hydrogen_bonding_1/3'>hydrogen bond network</scene> (<scene name='69/694230/Hydrogen_bonding_2/2'>with measurements</scene>) is represented by each atom type in the 3D model. The hydrogen bond network is characteristic of the MarR family as a whole. | ||
| - | [[Image:H Bonding of DNA.png|300 px|left|thumb|'''Figure 3''' The Hydrogen Bonding Network is shown with dotted green lines approximately 2.8 angstroms between residues. The network consists of 4 major residues as follows from right to left: E24, N38, Q40, S74.]] | + | [[Image:H Bonding of DNA.png|300 px|left|thumb|'''Figure 3'''. The Hydrogen Bonding Network is shown with dotted green lines approximately 2.8 angstroms between residues. The network consists of 4 major residues as follows from right to left: E24, N38, Q40, S74.]] |
== '''Zn(II) Binding''' == | == '''Zn(II) Binding''' == | ||
Revision as of 03:01, 19 April 2017
Adhesin Competence Regulator
| |||||||||||
References
- ↑ Sanson M, Makthal N, Flores AR, Olsen RJ, Musser JM, Kumaraswami M. Adhesin competence repressor (AdcR) from Streptococcus pyogenes controls adaptive responses to zinc limitation and contributes to virulence. Nucleic Acids Res. 2015 Jan;43(1):418-32. doi: 10.1093/nar/gku1304. Epub 2014 Dec, 15. PMID:25510500 doi:http://dx.doi.org/10.1093/nar/gku1304
- ↑ Fraústo da Silva J, Williams R. The Biological Chemistry of Elements: The Inorganic Chemistry of Life. Second ed. Oxford University Press; Oxford: 2001.
- ↑ Ma Z, Jacobsen FE, Giedroc DP. Coordination chemistry of bacterial metal transport and sensing. Chem Rev. 2009 Oct;109(10):4644-81. doi: 10.1021/cr900077w. PMID:19788177 doi:http://dx.doi.org/10.1021/cr900077w
- ↑ 4.0 4.1 4.2 4.3 4.4 Guerra AJ, Dann CE, Giedroc DP. Crystal Structure of the Zinc-Dependent MarR Family Transcriptional Regulator AdcR in the Zn(II)-Bound State. J Am Chem Soc. 2011 Nov 21. PMID:22085181 doi:10.1021/ja2080532
- ↑ 5.0 5.1 Reyes-Caballero H, Guerra AJ, Jacobsen FE, Kazmierczak KM, Cowart D, Koppolu UM, Scott RA, Winkler ME, Giedroc DP. The metalloregulatory zinc site in Streptococcus pneumoniae AdcR, a zinc-activated MarR family repressor. J Mol Biol. 2010 Oct 22;403(2):197-216. doi: 10.1016/j.jmb.2010.08.030. Epub 2010, Sep 8. PMID:20804771 doi:http://dx.doi.org/10.1016/j.jmb.2010.08.030