We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Luke Edward Severinac/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
In addition to a self-cleavage mechanism, Caspase-6 <scene name='75/752344/Caspase-6_zymogen_yeahboi/1'>zymogen</scene> can be activated getting cleaved by Caspase-3, as well as other enzymes. The mechanism of activation by clevage is highly conserved across the caspase family; Self-processing is uniquely recognized as the primary mechanism for Caspase-6 activation, where clevage must occur at <scene name='75/752344/Caspase-6_cleavage_sites_real/1'>three sites</scene> for complete activation, specifically the <scene name='75/752344/Caspase-6_prodomain/1'>pro-domain</scene> and the <scene name='75/752344/Caspase-6_intersubunit_linker/1'>intersubunit linker</scene> must be removed. These cleavages are both sequence specific and ordered, starting with cleavage of the pro-domain at <scene name='75/752344/Caspase-6_prodomain_cleavage/1'>residue 30</scene>. Then removal of the intersubunit linker occurs through cleavage at two sites, <scene name='75/752344/Caspase-6_176-179_cleavageyis/1'>DVVD179 and TEVD193</scene>. It has been proposed that this sequence of cleavage is due to the pro-domain being more readily available to enter the active site, whose presence inhibits Caspase-6's ability to cleave the intersubunit loop and self-activate; The prodomain acts as a “suicide protector”, preventing the TEVD193 cleavage site from the active site[3]. After both cleavages occur, <scene name='75/752344/Active_caspase_6_dimer/1'>active caspase-6</scene> remains in solution as a dimer. | In addition to a self-cleavage mechanism, Caspase-6 <scene name='75/752344/Caspase-6_zymogen_yeahboi/1'>zymogen</scene> can be activated getting cleaved by Caspase-3, as well as other enzymes. The mechanism of activation by clevage is highly conserved across the caspase family; Self-processing is uniquely recognized as the primary mechanism for Caspase-6 activation, where clevage must occur at <scene name='75/752344/Caspase-6_cleavage_sites_real/1'>three sites</scene> for complete activation, specifically the <scene name='75/752344/Caspase-6_prodomain/1'>pro-domain</scene> and the <scene name='75/752344/Caspase-6_intersubunit_linker/1'>intersubunit linker</scene> must be removed. These cleavages are both sequence specific and ordered, starting with cleavage of the pro-domain at <scene name='75/752344/Caspase-6_prodomain_cleavage/1'>residue 30</scene>. Then removal of the intersubunit linker occurs through cleavage at two sites, <scene name='75/752344/Caspase-6_176-179_cleavageyis/1'>DVVD179 and TEVD193</scene>. It has been proposed that this sequence of cleavage is due to the pro-domain being more readily available to enter the active site, whose presence inhibits Caspase-6's ability to cleave the intersubunit loop and self-activate; The prodomain acts as a “suicide protector”, preventing the TEVD193 cleavage site from the active site[3]. After both cleavages occur, <scene name='75/752344/Active_caspase_6_dimer/1'>active caspase-6</scene> remains in solution as a dimer. | ||
| - | ==Active State==[[Image:Binding grove active caspase 6.png|100 px|right|thumb|Substrate binding groove in Caspase-6. Blue - catalytic residues | + | ==Active State== |
| + | [[Image:Binding grove active caspase 6.png|100 px|right|thumb|Substrate binding groove in Caspase-6. Blue - catalytic residues | ||
yellow - ligand | yellow - ligand | ||
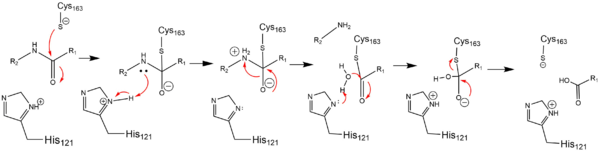
| - | red - generic surface]] | + | red - generic surface]]In order to function as an endoprotease, Caspase-6 utilzes a catalytic triad that binds a <scene name='75/752344/Protein_ligand_real/1'>ligand</scene>, which can include neuronal proteins and [https://en.wikipedia.org/wiki/Tubulin tubulins], in its active site. This binding groove contains three critical amino acid residues necessary to perform cleavage of the peptide bonds. A <scene name='75/752344/Catalytic_triad_real/1'>catalytic triad</scene>, composed of <scene name='75/752344/His121_real/1'>His-121</scene>, <scene name='75/752344/Glu123_real/1'>Glu-123</scene>, and <scene name='75/752344/Cys163_real/1'>Cys-163</scene>, carries out cleavage of the substrate. In the theorized mechanism, His-121 acts as an acid catalyst, Glu-123 acts as a base catalyst to deprotonate Cys-163, which then acts as covalent catalyst. |
| - | In order to function as an endoprotease, Caspase-6 utilzes a catalytic triad that binds a <scene name='75/752344/Protein_ligand_real/1'>ligand</scene>, which can include neuronal proteins and [https://en.wikipedia.org/wiki/Tubulin tubulins], in its active site. This binding groove contains three critical amino acid residues necessary to perform cleavage of the peptide bonds. A <scene name='75/752344/Catalytic_triad_real/1'>catalytic triad</scene>, composed of <scene name='75/752344/His121_real/1'>His-121</scene>, <scene name='75/752344/Glu123_real/1'>Glu-123</scene>, and <scene name='75/752344/Cys163_real/1'>Cys-163</scene>, carries out cleavage of the substrate. In the theorized mechanism, His-121 acts as an acid catalyst, Glu-123 acts as a base catalyst to deprotonate Cys-163, which then acts as covalent catalyst. | + | |
[[Image:Cystine Aspartase.png|600 px|active site mechanism]] | [[Image:Cystine Aspartase.png|600 px|active site mechanism]] | ||
Revision as of 03:15, 20 April 2017
Caspase-6 in Homo sapiens
| |||||||||||