Biological Function
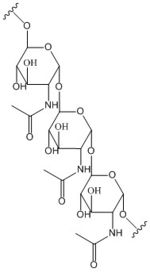
poly-β-1,6-N-acetylglucosamine
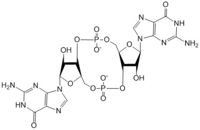
Diguanylate cyclases are a group of class 2 transferase enzymes that catalyze the production of cyclic dimeric-guanosine monophosphate (c-di-GMP), an important second messenger for signal transduction. Most commonly through phosphorylation or dephosphoylation events, signal transduction sends messages through cells to promote responses. Escherechia coli, a gram-negative bacterium often found in the intestines of mammals, uses diguanylate cyclase DgcZ in the synthesis of its biofilm. Enzyme DgcZ from E. coli acts as a catalyst to synthesize cyclic di-GMP from two substrate guanosine triphosphate (GTP) molecules to aid in communication of signals throughout the bacteria. C-di-GMP is a second messenger in the production of poly-β-1,6-N-acetylglucosamine (poly-GlcNAc), a polysaccharide required for E. coli biofilm production. This biofilm allows E. coli to adhere to extracellular surfaces. The enzyme is only successfully crystallized in its inactive conformation.

Diagram of DgcZ in its active (left) and inactive (right) conformations. Binding Zinc inactivates the enzyme.
Structural Overview

Diguanylate cyclase DgcZ from “E. Coli” with Domains Labeled
DgcZ is a tetrameric protein from E. coli made of two domains. Each domain is a dimer, so the whole protein can be called a dimer of dimers. The DgcZ protein has symmetry down its central axis. The catalytic glycine-glycine-glutamate-glutamate-phenylalanine (GGEEF) domain is responsible for synthesizing c-di-GMP, and the regulatory chemoreceptor zinc binding (CZB) domain houses two zinc binding sites. DgcZ binds zinc in the CZB domain with sub-femtomolar (10-16M) affinity. When zinc is bound, the CZB and GGEEF domains adopt conformations that inhibit DgcZ function [1]. Enzyme DgcZ was co-crystallized with Zinc fixing the structure in its inactivate conformation. The CZB domain is common to many bacterial lineages, including its prevalence in DgcZ homologs. The domain has an important role in signal transduction of bacteria. CZB and GGEEF domains are prevalent in many bacterial proteins from differing strands of E. coli [2]. The GGEEF domain is catalytic in that it contains the active sites used for cyclizing GTP into c-di-GMP. The CZB domain is used for ligand-mediated regulation of c-di-GMP production.
Catalytic GGEEF Domain
The domain of DgcZ is part of the GGDEF family of proteins that includes a conserved sequence, GG[DE][DE]F[3].The GGEEF domain is a homodimer consisting of a central five-stranded β-sheet surrounded by five α-helices. The GGEEF domain contains two catalytic half sites that, when combined together in a productive conformation, form the entire . Residues Gly-206, Gly-207, Glu-208, Glu-209, and Phe-210 form the active site. Each binds one GTP molecule. DgcZ binds the guanine base of GTP through hydrogen bonds to . This scene depicts an inactive form of the protein because it was crystallized with zinc bound. The ribose of each guanosine triphosphate, and subsequent product c-di-GMP riboses, are held only loosely by the enzyme, while the phosphate groups are not bound to the active site at all[1].
The alpha phosphate is available for attack by the 3 prime hydroxyl group on another GTP. A (Mg2+) stabilizes the negative charges on the phosphate groups. When in the productive conformation, each GTP is held in close proximity with the α-phosphate groups overlapping C3 of the ribose ring. This conformation allows the α-phospate of one GTP to react with the alcohol group attached to C3 of the ribose on the second GTP, resulting in a cyclization of the two molecules into c-di-GMP.
Mechanism of Action
Diguanylate cyclases only function efficiently as dimers, to bind both GGDEF domains holding the substrates. The presence of Zinc disrupts the ability of the two domains to overlap.
1. The enzyme coordinates the substrate GTP in a conformation to allow deprotonation of the C3 alcohol groups of the ribose. The negatively charged Oxygens on the phosphate groups of GTP are stabilized by Mg2+ ions.
2. The deprotonated oxygen then acts as a nucleophile to attack the α-phosphate of GTP. This initiates an addition-elimination reaction.
3. The β and γ phosphates of GTP are kicked off as leaving groups.
4. The result of this reaction is the C3 alcohol group of each ribose covalently bonded to an α-phosphate forming c-di-GMP.
CZB Domain and Zinc Binding Site
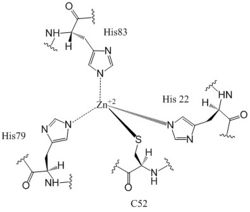
The , residues 19-90, is responsible for regulating the function of DgcZ due to the presence of two zinc binding sites. The domain contains the of the enzyme which exhibits cooperative binding. Four residues bind zinc with a high affinity even at 10-16M concentrations of Zinc in solution. Due to the tightness of Zinc binding, the complete enzyme has not yet been crystallized in its active conformation without the presence of Zinc metal inhibitor. When zinc is bound, DgcZ activity is limited[1]. Two Zinc binding sites are located on the CZB domain.
Zn
+2 Coordination to amino acid residues on three of the four 𝝰 helices of DgcZ
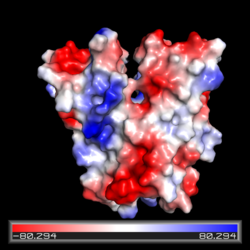
Electrostatic potential map of the CZB domain of Diguanylate Cyclase. Regions of relatively negative charge are in red and regions of relatively positive charge are in blue. Electrically neutral regions are in white
Most cells possess efficient Zinc uptake systems, as Zinc is a reactive Lewis Acid. Zinc binds incredibly tightly to this enzyme at subfemtomolar concentrations, attributing to why the enzyme has not been crystallized without Zinc present. The Zinc co-purified with the protein. Zinc allosterically inhibits the activity of enzyme DgcZ through two allosteric binding sites located on the CZB domain [1]. The inhibition prevents regulation of GGDEF domain function, the location of the active site. The CZB domain is folded into four anti-parallel α-helices as a 2-fold symmetric homodimer, with the N-terminus on the helix 𝝰4. The allosteric binding site includes a 3His/1Cys motif that uses amino acids H22 of 𝝰1, C52 of 𝝰2, and H79 and H83 of 𝝰3, spanning three of the four alpha helices of the CZB domain and coordinating the Zinc residue in a tetrahedral fashion. For clarification, the entirety of 𝝰helix 2 on one monomer of CZB is not successfully crystallized after the Cys52 residue and is not the N-terminal residue.
Zahringer et al. mutated Cys52 to Ala through site-directed mutagenesis, resulting in a lack of coordination on α2. The cysteine residue is not essential for Zinc binding, as Zinc still coordinates to the three His residues with the Cys52Ala mutation, but α2 is free to move and expose the Zinc binding pocket. This exposure was found to lower the protein's affinity for zinc, as the mutation of cysteine to alanine increased the activity of the DgcZ. Using EDTA, Zinc can be removed from the CZB domain. When not coordinated to zinc, the CZB domain presumably adopts a conformation that straightens the , shifting on the α-helices into the center and the GGEEF domain into its productive conformation, increasing activity of DgcZ. Activity increases without Zinc due to activation of poly-GlcNAc production and biofilm formation, and maximal cyclic di-GMP production.
Other Ligands
c-di-GMP and GTP bind although the function of this binding is unknown. Very weak product inhibition was observed when c-di-GMP bound allosterically but the inhibition was so weak, it is possible the c-di-GMP actually interacts with another as of yet unknown molecule at that site.
References
1. Zahringer, Franziska, Egidio Lancanna, Urs Jenal, Tilman Schirmer, and Alex Boehm. "Structure of E. Coli Zinc-Sensory Diguanylate Cyclase DgcZ." Cell Press: Structure 21 (2013): 1149-157.
2. Jenny Draper, K. Karplus, K. Ottemann. Identification of a Chemoreceptor Zinc-Binding Domain Common to Cytoplasmic Bacterial Chemoreceptors. Journal of Bacteriology. Vol. 193, No. 17. 4338-4345. (2011).
3. Carmen Chan, R. Paul, D. Samoray, N. Amiot, B. Giese, U. Jenal, T. Schirmer. Structural basis of activity and allosteric control of diguanylate cyclases. PNAS. Vol 101. No. 49 17084-17089. (2004).