We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1072
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
== Biological Function == | == Biological Function == | ||
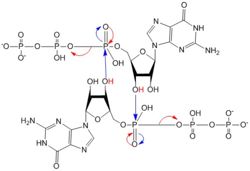
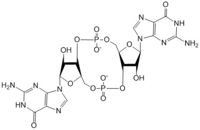
[[Image:C-di-GMP larger font.jpg |200 px|thumb|left|'''Figure 1: Cyclic-dimeric-GMP.''' Cyclic-dimeric-GMP is the product of the reaction catalyzed by DgcZ.]] | [[Image:C-di-GMP larger font.jpg |200 px|thumb|left|'''Figure 1: Cyclic-dimeric-GMP.''' Cyclic-dimeric-GMP is the product of the reaction catalyzed by DgcZ.]] | ||
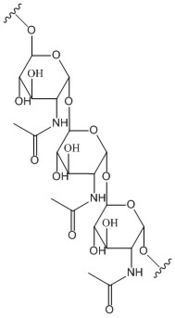
| - | [[Image:Poly B-1, 6 GlcNAc.jpg | | + | [[Image:Poly B-1, 6 GlcNAc.jpg |175 px|left|thumb|'''Figure 2: Poly-β-1,6-N-acetylglucosamine.''' Poly-β-1,6-N-acetylglucosamine is a carbohydrate formed downstream of c-di-GMP utilized in formation of bacterial bio-films.]] |
Diguanylate cyclases are class 2 transferase enzymes <span class="plainlinks">[https://en.wikipedia.org/wiki/Diguanylate_cyclase (2.7.7.65)]</span> that catalyze the production of cyclic dimeric-guanosine monophosphate (c-di-GMP, Figure 1), important to <span class="plainlinks">[https://en.wikipedia.org/wiki/Signal_transduction signal transduction]</span> as a <span class="plainlinks">[https://en.wikipedia.org/wiki/Second_messenger_system second messenger]</span><sup>[1]</sup>. Signal transduction sends messages through cells to induce cellular responses, most commonly through phosphorylation or dephosphoylation of substrate molecules. <span class="plainlinks">[https://en.wikipedia.org/wiki/Escherichia_coli ''Escherechia coli (E. coli)'']</span>, a gram-negative bacterium often found in the intestines of mammals, uses diguanylate cyclase (DgcZ) in the synthesis of its <span class="plainlinks">[https://en.wikipedia.org/wiki/biofilm biofilm]</span><sup>[2]</sup>. DgcZ from ''E. coli'' acts as a catalyst, synthesizing c-di-GMP from two substrate guanosine triphosphate (GTP) molecules. C-di-GMP is a second messenger in the production of poly-β-1,6-N-acetylglucosamine (poly-GlcNAc, Figure 2), a polysaccharide required for ''E. coli'' biofilm production<sup>[2]</sup>. This biofilm allows ''E. coli'' to adhere to extracellular surfaces. Only the inactive conformation of the complete DgcZ enzyme has been crystallized to date<sup>[3]</sup>. | Diguanylate cyclases are class 2 transferase enzymes <span class="plainlinks">[https://en.wikipedia.org/wiki/Diguanylate_cyclase (2.7.7.65)]</span> that catalyze the production of cyclic dimeric-guanosine monophosphate (c-di-GMP, Figure 1), important to <span class="plainlinks">[https://en.wikipedia.org/wiki/Signal_transduction signal transduction]</span> as a <span class="plainlinks">[https://en.wikipedia.org/wiki/Second_messenger_system second messenger]</span><sup>[1]</sup>. Signal transduction sends messages through cells to induce cellular responses, most commonly through phosphorylation or dephosphoylation of substrate molecules. <span class="plainlinks">[https://en.wikipedia.org/wiki/Escherichia_coli ''Escherechia coli (E. coli)'']</span>, a gram-negative bacterium often found in the intestines of mammals, uses diguanylate cyclase (DgcZ) in the synthesis of its <span class="plainlinks">[https://en.wikipedia.org/wiki/biofilm biofilm]</span><sup>[2]</sup>. DgcZ from ''E. coli'' acts as a catalyst, synthesizing c-di-GMP from two substrate guanosine triphosphate (GTP) molecules. C-di-GMP is a second messenger in the production of poly-β-1,6-N-acetylglucosamine (poly-GlcNAc, Figure 2), a polysaccharide required for ''E. coli'' biofilm production<sup>[2]</sup>. This biofilm allows ''E. coli'' to adhere to extracellular surfaces. Only the inactive conformation of the complete DgcZ enzyme has been crystallized to date<sup>[3]</sup>. | ||
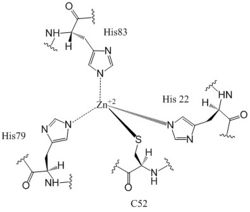
[[Image:Conformation change 2 bold.png|250 px|right|thumb|'''Figure 3: Diagram of DgcZ.''' DgcZ is shown in its active (left) and inactive (right) conformations. The boxes represent the GGEEF domains of the enzyme, while the cylinders represent the alpha helices of the CZB domains where the Zinc binding sites are located<sup>[3]</sup>. Binding Zn<sup>+2</sup> inactivates the enzyme, disrupting the active conformation of the GGEEF domain by not allowing the GTP molecules to react. The red and blue sections represent the two monomers of the symmetric homodimer.]] | [[Image:Conformation change 2 bold.png|250 px|right|thumb|'''Figure 3: Diagram of DgcZ.''' DgcZ is shown in its active (left) and inactive (right) conformations. The boxes represent the GGEEF domains of the enzyme, while the cylinders represent the alpha helices of the CZB domains where the Zinc binding sites are located<sup>[3]</sup>. Binding Zn<sup>+2</sup> inactivates the enzyme, disrupting the active conformation of the GGEEF domain by not allowing the GTP molecules to react. The red and blue sections represent the two monomers of the symmetric homodimer.]] | ||
| Line 18: | Line 18: | ||
===Mechanism of Action=== | ===Mechanism of Action=== | ||
| - | [[Image:C-di-GMP arrows.jpg|250 px|right|thumb|'''Figure 5: Mechanism of c-di-GMP synthesis.''' Addition arrows are shown in blue and elimination arrows are shown in red.]] Diguanylate cyclases only function efficiently when the two half-sites of the GGEEF | + | [[Image:C-di-GMP arrows.jpg|250 px|right|thumb|'''Figure 5: Mechanism of c-di-GMP synthesis.''' Addition arrows are shown in blue and elimination arrows are shown in red.]] Diguanylate cyclases only function efficiently when the two half-sites of the GGEEF domains are in close enough proximity to bind the substrates. The presence of Zinc disrupts the ability of the domains to overlap. |
1. The enzyme coordinates the substrate <scene name='69/694239/Gtp_alone/5'>GTP</scene> in a conformation to allow deprotonation of the C3 alcohol groups of the ribose. The negatively charged Oxygens on the phosphate groups of GTP are stabilized by Mg<sup>2+</sup> ions. | 1. The enzyme coordinates the substrate <scene name='69/694239/Gtp_alone/5'>GTP</scene> in a conformation to allow deprotonation of the C3 alcohol groups of the ribose. The negatively charged Oxygens on the phosphate groups of GTP are stabilized by Mg<sup>2+</sup> ions. | ||
Revision as of 02:51, 22 April 2017
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Diguanylate Cyclase DgcZ from Escherichia coli
| |||||||||||
![Figure 3: Diagram of DgcZ. DgcZ is shown in its active (left) and inactive (right) conformations. The boxes represent the GGEEF domains of the enzyme, while the cylinders represent the alpha helices of the CZB domains where the Zinc binding sites are located[3]. Binding Zn+2 inactivates the enzyme, disrupting the active conformation of the GGEEF domain by not allowing the GTP molecules to react. The red and blue sections represent the two monomers of the symmetric homodimer.](/wiki/images/thumb/1/10/Conformation_change_2_bold.png/250px-Conformation_change_2_bold.png)