User:Luke Edward Severinac/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
<StructureSection load='4IYR' size='340' side=http://proteopedia.org/wiki/index.php?title=User:Luke_Edward_Severinac/Sandbox_1&action=edit'right' caption='Caspase-6' scene=''> | <StructureSection load='4IYR' size='340' side=http://proteopedia.org/wiki/index.php?title=User:Luke_Edward_Severinac/Sandbox_1&action=edit'right' caption='Caspase-6' scene=''> | ||
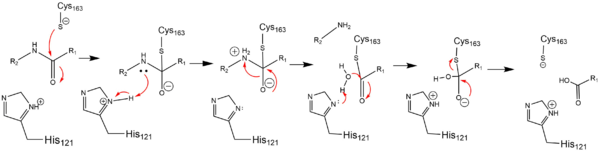
| - | Caspase-6 is an [https://en.wikipedia.org/wiki/Endopeptidase endopeptidase] involved in apoptosis. In terms of its catalytic function, it is a part of the [https://en.wikipedia.org/wiki/Caspase cysteine-aspartate family]. Before Caspase-6 becomes functional, the enzyme exists as a <scene name='75/752344/Caspase-6_zymogen/1'>procaspase</scene>, also known as a [https://en.wikipedia.org/wiki/Zymogen zymogen]. This zymogen exists as a <scene name='75/752344/Caspase-6_zymogen/1'>homodimer</scene>, whose <scene name='75/752344/Caspase-6_zymogen_realller/1'>monomeric units</scene> are then cleaved at <scene name='75/752344/Caspase-6_cleavage_sites_real/1'>specific sites</scene> to assume its active conformation. Zymogen activation through cleavage is largely conserved across the caspase family. However, Caspase-6 is unique in that it becomes active through self-cleavage in addition to cleavage by a separate enzymes. Each monomeric unit of zymogen contains a <scene name='75/752344/Caspase-6_small_subunit_mnmr/1'>small subunit</scene> consisting of two helices, a <scene name='75/752344/Caspase-6_large_real_yeahboi/1'>large subunit</scene> consisting of three helices, a <scene name='75/752344/Caspase-6_prodomain/1'>prodomain</scene>, and a <scene name='75/752344/Caspase-6_zymogen_b-sheet/1'>beta sheet core</scene>. After cleavage at all sites, the processed post-zymogen monomers remain closely associated together through intermolecular forces as a dimer. | + | Caspase-6 is an [https://en.wikipedia.org/wiki/Endopeptidase endopeptidase] involved in apoptosis. In terms of its catalytic function, it is a part of the [https://en.wikipedia.org/wiki/Caspase cysteine-aspartate family]. Before Caspase-6 becomes functional, the enzyme exists as a <scene name='75/752344/Caspase-6_zymogen/1'>procaspase</scene>, also known as a [https://en.wikipedia.org/wiki/Zymogen zymogen]. This zymogen exists as a <scene name='75/752344/Caspase-6_zymogen/1'>homodimer</scene>, whose <scene name='75/752344/Caspase-6_zymogen_realller/1'>monomeric units</scene> are then cleaved at <scene name='75/752344/Caspase-6_cleavage_sites_real/1'>specific sites</scene> to assume its active conformation. Zymogen activation through cleavage is largely conserved across the caspase family. However, Caspase-6 is unique in that it becomes active through self-cleavage in addition to cleavage by a separate enzymes<ref name="zincmedallinhib">PMID: 22891250 </ref>. Each monomeric unit of zymogen contains a <scene name='75/752344/Caspase-6_small_subunit_mnmr/1'>small subunit</scene> consisting of two helices, a <scene name='75/752344/Caspase-6_large_real_yeahboi/1'>large subunit</scene> consisting of three helices, a <scene name='75/752344/Caspase-6_prodomain/1'>prodomain</scene>, and a <scene name='75/752344/Caspase-6_zymogen_b-sheet/1'>beta sheet core</scene>. After cleavage at all sites, the processed post-zymogen monomers remain closely associated together through intermolecular forces as a dimer. |
=='''Zymogen'''== | =='''Zymogen'''== | ||
| Line 21: | Line 21: | ||
=='''Phosphorylation'''== | =='''Phosphorylation'''== | ||
| - | The function of Caspase-6 can be inhibited by phosphorylation of Ser-257. The exact mechanism of this reaction remains unidentified at the time of publication, but proceeds when ARK5 kinase is present. This modification can occur before and after zymogen activation. The phosphoryl group inhibits Caspase-6 through steric interference. When Ser-257 is phosphorylated, the amino acid residue interacts with <scene name='75/752344/Caspase-6_his-208/1'>Pro-201</scene>, causing a shift in the helices of Caspase-6. This is shown in the <scene name='75/752344/Caspase-6_s257d_mutant/1'>S257D Caspase-6 mutant</scene>, whose mutation mimics phosphorylation. The shift misaligns and disrupts residues found in the active site. This conformational difference prevents the intersubunit linker from entering during zymogen activation and the self-cleaved active dimer cannot be formed. Additionally, no new substrate is able to enter the active site. | + | The function of Caspase-6 can be inhibited by phosphorylation of Ser-257. The exact mechanism of this reaction remains unidentified at the time of publication, but proceeds when ARK5 kinase is present. This modification can occur before and after zymogen activation. The phosphoryl group inhibits Caspase-6 through steric interference. When Ser-257 is phosphorylated, the amino acid residue interacts with <scene name='75/752344/Caspase-6_his-208/1'>Pro-201</scene>, causing a shift in the helices of Caspase-6. This is shown in the <scene name='75/752344/Caspase-6_s257d_mutant/1'>S257D Caspase-6 mutant</scene>, whose mutation mimics phosphorylation<ref name="Phosregcasp6subsbindgroove">PMID: 22483120 </ref>. The shift misaligns and disrupts residues found in the active site. This conformational difference prevents the intersubunit linker from entering during zymogen activation and the self-cleaved active dimer cannot be formed. Additionally, no new substrate is able to enter the active site. |
=='''Medical Relevance'''== | =='''Medical Relevance'''== | ||
| Line 28: | Line 28: | ||
=='''References'''== | =='''References'''== | ||
| + | {{reflist}} | ||
Revision as of 02:17, 23 April 2017
Caspase-6 in Homo sapiens
| |||||||||||