We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1236
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
{{Sandbox_Reserved_Jason_Telford}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | {{Sandbox_Reserved_Jason_Telford}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
==Luciferase== | ==Luciferase== | ||
| - | <StructureSection load='5KYV'> | + | <StructureSection load='5KYV'> |
| + | |||
| + | Luciferase is an enzyme responsible for catalyzation of many bioluminencent pathways found in various organism. It is a highly studied and utilized protein for applications in cell biology, genetics and biochemistry. | ||
== Function == | == Function == | ||
| Line 19: | Line 21: | ||
== Relevance == | == Relevance == | ||
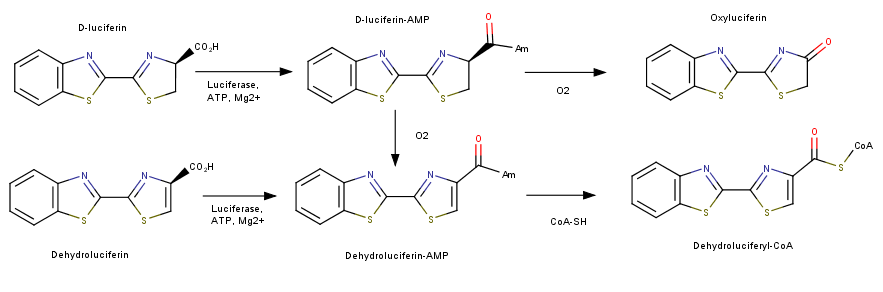
| - | This protein has been utilized in various types of assays ranging from quantification of ATP and the rate of transcription within a cell.<ref>Wet, J. R., Wood, K. V., Helinski, D. R., & Deluca, M. (1985). Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli. Proceedings of the National Academy of Sciences, 82(23), 7870-7873. </ref>. This molecule is especially unique due to the fact that is very efficient in producing a photon through this reaction. Luciferase is sensitive to small changes in substrate and is a optimal choice for quantification of gene expression. It has potential for further biological applications in the future. Luciferase is widely used as a luminescent reporter gene in a variety of assays. The Luciferase gene can be isolated from the firefly and be | + | This protein has been utilized in various types of assays ranging from quantification of ATP and the rate of transcription within a cell.<ref>Wet, J. R., Wood, K. V., Helinski, D. R., & Deluca, M. (1985). Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli. Proceedings of the National Academy of Sciences, 82(23), 7870-7873. </ref>. This molecule is especially unique due to the fact that is very efficient in producing a photon through this reaction. Luciferase is sensitive to small changes in substrate and is a optimal choice for quantification of gene expression. It has potential for further biological applications in the future. Luciferase is widely used as a luminescent reporter gene in a variety of assays. The Luciferase gene can be isolated from the firefly and be insterted into a plasmid. This plasmid could contain a gene that is destined for transfection into a cell. After transfection luciferin can be added to a cell culture in order to visualize the expression of the transfected gene. The amount of fluorescence can be quantified to determine the amount of expression within a cell. <ref>"Luciferase Assays." Thermo Fisher Scientific. Thermo Fisher Scientific, 2017. Web. 28 Apr. 2017.</ref> |
== Structural highlights == | == Structural highlights == | ||
| - | The structure of this protein comprises of two prominent domains. The larger one contains an N terminal distorted beta-barrel accompanied by alpha helices. The second and smaller unit is consist of a beta sheet and alpha helix complex <ref>Viviani, V. R. (2002). The origin, diversity, and structure function relationships of insect luciferases. Cellular and Molecular Life Sciences, 59(11), 1833-1850. </ref>. The process of fluorescence is achieved through a two-step oxidation reaction involving the substrate Lucinferin accompanied with ATP, Magnesium and oxygen. The first step consist of using ATP-Mg in an Acylation reaction of the COOH group on Lucinferin producing a Luciferyl adenylate intermediate and a phosphate group. The second reaction uses oxygen to create an excited state of the molecule. The molecule then returns to its ground state emitting a photon of light | + | The structure of this protein comprises of two prominent domains. The larger one contains an N terminal distorted beta-barrel accompanied by alpha helices. The second and smaller unit is consist of a beta sheet and alpha helix complex <ref>Viviani, V. R. (2002). The origin, diversity, and structure function relationships of insect luciferases. Cellular and Molecular Life Sciences, 59(11), 1833-1850. </ref>. The process of fluorescence is achieved through a two-step oxidation reaction involving the substrate Lucinferin accompanied with ATP, Magnesium and oxygen. The first step consist of using ATP-Mg in an Acylation reaction of the COOH group on Lucinferin producing a Luciferyl adenylate intermediate and a phosphate group. The second reaction uses oxygen to create an excited state of the molecule. The molecule then returns to its ground state emitting a photon of light. <ref>Conti, E., Franks, N. P., & Brick, P. (1996). Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure, 4(3), 287-298.</ref> |
| Line 33: | Line 35: | ||
==Inhibitors== | ==Inhibitors== | ||
Inhibition of luciferase can be mediated by several types of molecules. The presence of CoA has been found to have down regulate the bioluminescencent pathway of luciferase. Increased CoA favors the nonluminescent pathway which does not create a photon of light and instead produces Dehydroluciferyl-CoA. In addition to CoA inhibition of this enzyme has been seen with the exposure to diverse group of anesthetics. These molecules bind allosterically to luciferase and cause a conformation change in the protein. This unfolding alters the active site of the protein enough to not allow the bioluminescent catalysis to occur. Other than macro molecules, certain metal ions, specifically Ni and Co, have been seen to inhibit this reaction by replacing Magnesium in the active site. Magnesium is required for use of ATP and without it the reaction will not take place. Of the many inhibitors identified, lipoic acid demonstrates strong activity. Lipoic acid competes with luciferin for the active site enabling the binding of the substrate for the reaction. <ref>Leitão, João M. M., and Joaquim C G Esteves Da Silva. "Firefly luciferase inhibition." Journal of photochemistry and photobiology. (2010): 1-8. Research Gate. Web. 28 Apr. 2017.</ref> | Inhibition of luciferase can be mediated by several types of molecules. The presence of CoA has been found to have down regulate the bioluminescencent pathway of luciferase. Increased CoA favors the nonluminescent pathway which does not create a photon of light and instead produces Dehydroluciferyl-CoA. In addition to CoA inhibition of this enzyme has been seen with the exposure to diverse group of anesthetics. These molecules bind allosterically to luciferase and cause a conformation change in the protein. This unfolding alters the active site of the protein enough to not allow the bioluminescent catalysis to occur. Other than macro molecules, certain metal ions, specifically Ni and Co, have been seen to inhibit this reaction by replacing Magnesium in the active site. Magnesium is required for use of ATP and without it the reaction will not take place. Of the many inhibitors identified, lipoic acid demonstrates strong activity. Lipoic acid competes with luciferin for the active site enabling the binding of the substrate for the reaction. <ref>Leitão, João M. M., and Joaquim C G Esteves Da Silva. "Firefly luciferase inhibition." Journal of photochemistry and photobiology. (2010): 1-8. Research Gate. Web. 28 Apr. 2017.</ref> | ||
| + | |||
| + | |||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 22:41, 28 April 2017
| This Sandbox is Reserved from Jan 17 through June 31, 2017 for use in the course Biochemistry II taught by Jason Telford at the Maryville University, St. Louis, USA. This reservation includes Sandbox Reserved 1225 through Sandbox Reserved 1244. |
To get started:
More help: Help:Editing |
Luciferase
| |||||||||||