Ribose-5-phosphate isomerase
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
==Structure== | ==Structure== | ||
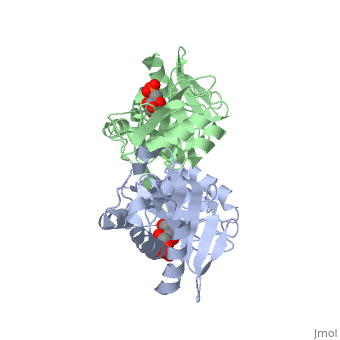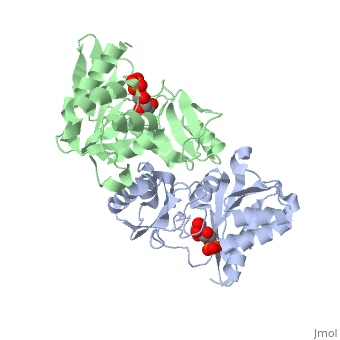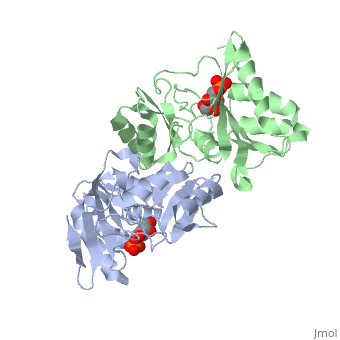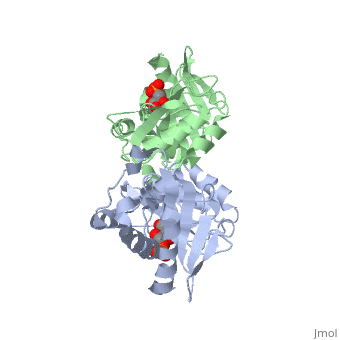
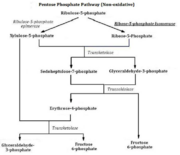
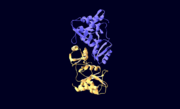
| - | The structure of RpiA has been identified in many organisms including ''E. coli'' and ''Vibrio vulnificus''. The crystallized RpiA structure from both of these organisms is highly conserved in many respects. RpiA exists as a dimer with pseudo-2-fold symmetry, the interface of the dimer is composed of six different segments, which contain a number of interactions occurring between <scene name='Sandbox_Reserved_305/Alpha_helices/2'> α-helices </scene> and <scene name='Sandbox_Reserved_305/Beta_sheets/1'> β-sheets </scene>. Two salt bridges also link <scene name=' | + | The structure of RpiA has been identified in many organisms including ''E. coli'' and ''Vibrio vulnificus''. The crystallized RpiA structure from both of these organisms is highly conserved in many respects. RpiA exists as a dimer with pseudo-2-fold symmetry, the interface of the dimer is composed of six different segments, which contain a number of interactions occurring between <scene name='Sandbox_Reserved_305/Alpha_helices/2'> α-helices </scene> and <scene name='Sandbox_Reserved_305/Beta_sheets/1'> β-sheets </scene>. Two salt bridges also link <scene name='44/446270/Cv/1'>Lys104 and Glu183</scene> of the same subunit. The <scene name='Sandbox_Reserved_305/Activesite/3'>RpiA and R5P complex</scene> occurs through the interaction between the ligand (R5P) and the following residues in the RpiA: <scene name='Sandbox_Reserved_305/Test/1'>Gly97, Asp84</scene>, Lys121, Lys7, Thr31 and Ser30. RpiA contains two sites for this interaction allowing two R5Ps to interact with one RpiA. The VvRpiA-R5P complex resembles the ''E. coli'' RpiA-A5P complex; however the VvRpiA-A5P complex reveals a different position than the R5P binding mode. The A5P interacts with the following residues: Asp8, Lys7, Ser30, Asp118 and Lys121. |
==Medical and Future Implications== | ==Medical and Future Implications== | ||
Revision as of 09:35, 14 May 2017
| |||||||||||
3D structures of ribose-5-phosphate isomerase
Updated on 14-May-2017
References
- ↑ 1.0 1.1 1.2 1.3 Zhang R, Andersson CE, Savchenko A, Skarina T, Evdokimova E, Beasley S, Arrowsmith CH, Edwards AM, Joachimiak A, Mowbray SL. Structure of Escherichia coli ribose-5-phosphate isomerase: a ubiquitous enzyme of the pentose phosphate pathway and the Calvin cycle. Structure. 2003 Jan;11(1):31-42. PMID:12517338
- ↑ 2.0 2.1 Kim TG, Kwon TH, Min K, Dong MS, Park YI, Ban C. Crystal structures of substrate and inhibitor complexes of ribose 5-phosphate isomerase A from Vibrio vulnificus YJ016. Mol Cells. 2009 Jan;27(1):99-103. Epub 2009 Feb 5. PMID:19214439 doi:10.1007/s10059-009-0010-6
- ↑ 3.0 3.1 3.2 Huck JH, Verhoeven NM, Struys EA, Salomons GS, Jakobs C, van der Knaap MS. Ribose-5-phosphate isomerase deficiency: new inborn error in the pentose phosphate pathway associated with a slowly progressive leukoencephalopathy. Am J Hum Genet. 2004 Apr;74(4):745-51. Epub 2004 Feb 25. PMID:14988808 doi:10.1086/383204
- ↑ 4.0 4.1 Gengenbacher M, Fitzpatrick TB, Raschle T, Flicker K, Sinning I, Muller S, Macheroux P, Tews I, Kappes B. Vitamin B6 biosynthesis by the malaria parasite Plasmodium falciparum: biochemical and structural insights. J Biol Chem. 2006 Feb 10;281(6):3633-41. Epub 2005 Dec 8. PMID:16339145 doi:10.1074/jbc.M508696200
- ↑ Salyers AA, Amabile-Cuevas CF. Why are antibiotic resistance genes so resistant to elimination? Antimicrob Agents Chemother. 1997 Nov;41(11):2321-5. PMID:9371327
- ↑ Kim TG, Kwon TH, Min K, Dong MS, Park YI, Ban C. Crystal structures of substrate and inhibitor complexes of ribose 5-phosphate isomerase A from Vibrio vulnificus YJ016. Mol Cells. 2009 Jan;27(1):99-103. Epub 2009 Feb 5. PMID:19214439 doi:10.1007/s10059-009-0010-6
- Created with the participation of Jasmeet Bhullar.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Andrea Gorrell, Jaime Prilusky