Structure and Function
Glycogen phosphorylase is a dimer consisting of two identical subunits and has an essential cofactor, [3]. Glycogen phosphorylase can be found in two different states, glycogen phosphorylase a (GPa) and glycogen phosphorylase b (GPb)[1]The difference in the structures is due to phosphorylation of the residue which results in the active form (GPa). Protein phosphatases dephosphorylate the GPa to the inactive form, also known as GPb. Both forms of glycogen phosphorylase can also be found in T and R states where T is the inactive state because it appears to have a low affinity for substrate and R is the active state where it appears to have a greater affinity for substrate[4].
The secondary structures of T and R states of GPa and b are similar with an and a .Each domain also contains subdomains which undergo conformational changes on the interconversion of T and R states [4]. C terminal domain has the cofactor PLP and part of the active site, it is made up of five α helices and 6 β strands[5]. The N-terminal domain consisting of fifteen α helices and nine β strands, is considered to be more complex and is divided in the middle of its β sheet core into subdomains. The first domain binds the effector molecule of AMP and also has a recognition site of the introconverting phosphorylase kinase and phosphatase[4][5]. The second domain has the polysaccharide binding domain where phosphorylase is able to attach to the glycogen substrate [5]. The R states of GPa and GPb are almost identical; the difference lays in the modification of the Ser-14 residue where GPa has a covalently linked phosphate group whereas GPb has a non-covalently linked sulfide group . GPa is activated by phosphorylation of the serine residue whereas GPb can be activated by the binding of AMP to the that are present within the molecule[4]. GPa does not require the binding of AMP but attachment enhances the activity of the enzyme upwards to 25% [6].
Glycogen phosphorylase is different from other enzymes that require the cofactor PLP because instead of utilizing the pyrimidine ring, phosphorylase uses the phosphate group [6]. The 4'aldehyde of PLP binds to the ε-amino group of lysine 680 and the 5'-phosphate of PLP has been found to be the group participating in the catalysis of glycogen phosphorylase [3]. The binding sites in glycogen phosphorylase include: a catalytic, inhibiting, AMP, glycogen and new allosteric site [1]. The glycogen binding site is located more than 30Å from the catalytic and allosteric sites. The residues that make up the site are [6]. The binds purine analogs or fused-ring molecules such as adenosine, caffeine, FMN, NADH and AMP when there are increased concentrations available[4]. The heterocyclic rings of the compounds bind to the inhibiting site, stablilizing it and blocking access to the catalytic center [6]. The can be accessed once the Ser-14 residue has been phosphorylated and conformational changes in glycogen phosphorylation have been observed[5][6]. The structure and function of glycogen phosphorylase is complex, though the function of the enzyme is due to the structure.
Mechanism
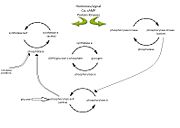
Figure 1: Diagram illustrating the enzyme cascade that occurs when hormones(and neural signals)catalyze the activation of phosphorylase through kinase activity and thus inactivating glycogen synthetase. Phosphatases inactivate phosphorylase and is regulated by phosphorylase itself as well as inhibitor proteins. High glucose concentratios also lead to the inactivation of phosphorylase (inactivation represented by open arrows). Diagram adapted from
[5].
In muscle, glycogen phosphorylase is activated by hormones and neural signals such as epinephrine, that stimulate phosphorylase kinase which phosphorylates the Ser-14 residue of the protein. A second messenger of cyclic AMP (cAMP) increases in concentration due to epinephrine or glucagon, and this increase results in an enzyme cascade
[5]. Activation of phosphorylase kinase is due to increased concentrations of Ca
2+ or by the phosphorylation by protein kinase A which is cAMP dependent. The activated kinase in turn activates the glycogen phosphorylase enzyme by phosphorylating the Ser-14 residue. In the liver, glucagon is the primary signal which catalyzes this enzyme cascade
[5][6].
Glycogen phosphorylase is regulated by phosphorylation, binding of allosteric effectors and by the catalytic mechanism; phosphorylation takes glycogen phosphorylase from a disordered state to an ordered one, allosteric effector provide changes in the structure of the enzyme and when coupled with phosphorylation allow access to the buried catalytic site[6]. The catalytic mechanism itself is dependent upon the proximity of PLP and the substrate phosphate which is directed by the surrounding groups which stabilize the interactions[5] and create the perfect environment to phosphohydrolyze the glycosidic bond[6]. The environment is established by the phosphate compound making a hydrogen bond with the 5'-phosphate of PLP and being stable enough to successfully cleave the bond yielding the product of glucose-1-phosphate.[3].
Reaction
Glycogen phosphorylase (GP) catalyzes the degradation of the reducing end of glycogen into glucose-1-phosphate. It employs a cofactor called pyridoxal-5’ –phosphate, that is located in the active site and bound to a K681 residue with a Schiff base linkage. PLP shuttles the phosphate group onto the substrate (written by Jaime Prilusky, Max Lein, Eran Hodis).
History
This protein comes from the muscle tissue of Oryctolagus cuniculus. There is an isozyme from liver tissue that is regulated by glucagon instead of epinephrine, with a different gene that encodes it and different regulation properties (written by Jaime Prilusky, Max Lein, Eran Hodis).
Glycogen phosphorylase was the first phosphorylase enzyme to be discovered, and the first example of regulation via covalent modification.
In the 1930s, the first work done by Carl and Gerty Cori. They proved that the enzyme exists in 'A' and 'B' forms, and they showed that the reverse reaction produced glycogen. They won the Nobel Prize in 1947 along with Bernardo Housay of Argentina for their work on carbohydrate metabolism. This was also the first example of a polymerizing enzyme, inspiring others to look for other polymerizing enzymes.
Subsequently, Earl Sutherland found that the 'B' form predominates in resting muscle and epinephrine triggers activation to form 'A'. Since then, many groups have worked on this enzyme, both to understand its mechanism and to discover drug targets. Crystal structures have been obtained for the protein in the 'A' and 'B' form, in the presence of natural substrates, inhibitors, and transition state analogs. Please see the end of this article for links to crystallographic information.
Activity and Regulation of GP
In its active form, GP is a dimer of two identical subunits. The subunits make interactions that stabilize the final structure.
Each Sub-unit contains 5 potential effector sites:
1. Ser14 phosphate-recognition site.
2. AMP activation / Glc-6-P inhibition site.
3. Catalytic site that binds glycogen, Glc-1-P
4. Inhibitor site, 12Å from catalytic site, binds caffeine and related compounds.
5. Glycogen storage site.
There are two forms of the enzyme, designated as 'A' and 'B', that are controlled hormonally. The 'B' form is converted into the 'A' form by phosphorylase kinase, which catalyzes the addition of phosphate from ATP to Ser14 near the N-terminus. This represents the final step in a signal transduction cascade in response to the hormone epinephrine, associated with the 'fight-or-flight' response and causing an increase in available energy to the organism as a whole. The N-terminus contains a high percentage of basic residues, which interact favorably with a pocket of acidic residues (Asp109, Glu110, Glu120, Glu501, Glu505 and Glu509) in the 'B' form. Once Ser14 is phosphorylated, the N-terminus is forced ~50Å away from the acidic residues, settling into a region with R69 and R45' (prime denotes a residue from the adjacent subunit). In summary, the conformatino change causes an ordering of the N-terminal chain and a disordering of residues at the C-terminal. Once disordered, the C-terminal residues are no longer able to block substrate entry into the active site. The enzyme phosphatase is able to remove the phosphate and return GP to form 'B'.
In addition, the 'A' and 'B' forms can be regulated futher by small molecules in the cell. This allows individual cells to ignore the hormonal signal if they already have enough available energy (at high concentrations of glucose derivatives or ATP, designated as the 'T' state for low substrate affinity), or activate GP without a hormonal signal when energy for the individual cell is needed (high concentrations of AMP, designated as the 'R' state for high substrate affinity; written by Jaime Prilusky, Max Lein, Eran Hodis).
References to the last 3 sections
- Barford D, Hu SH, Johnson LN. Structural mechanism for glycogen phosphorylase control by phosphorylation and AMP. J Mol Biol. 1991 Mar 5;218(1):233-60. PMID:1900534
- Oikonomakos NG, Skamnaki VT, Tsitsanou KE, Gavalas NG, Johnson LN. A new allosteric site in glycogen phosphorylase b as a target for drug interactions. Structure. 2000 Jun 15;8(6):575-84. PMID:10873856
- Johnson LN, Barford D. Electrostatic effects in the control of glycogen phosphorylase by phosphorylation. Protein Sci. 1994 Oct;3(10):1726-30. PMID:7849589
Links
1GPA is a Single protein structure of sequence from Oryctolagus cuniculus. Additional information on 1GPA is available in a page on Glycogen Phosphorylase at the RCSB PDB Molecule of the Month. Full crystallographic information is available from OCA.