Hyaluronidase
From Proteopedia
(Difference between revisions)
| Line 36: | Line 36: | ||
**[[2jnk]] - CpHU residues 1498-1628]] - NMR<br /> | **[[2jnk]] - CpHU residues 1498-1628]] - NMR<br /> | ||
**[[2atm]] - HU – ''Vespula vulgaris''<br /> | **[[2atm]] - HU – ''Vespula vulgaris''<br /> | ||
| - | **[[2pe4]] – HU-1 – human | + | **[[2pe4]] – HU-1 – human<br /> |
| + | **[[4ufq]] – HU – ''Streptomyces koganeiensis''<br /> | ||
*Hyaluronidase binary complex | *Hyaluronidase binary complex | ||
| Line 60: | Line 61: | ||
**[[2yvv]] - SpyHU + lactose<br /> | **[[2yvv]] - SpyHU + lactose<br /> | ||
**[[3eka]] - SpyHU + ascorbate <br /> | **[[3eka]] - SpyHU + ascorbate <br /> | ||
| + | **[[5diy]] – HU + Map3K 7 interacting protein peptide – ''Thermobaculum terrenum''<br /> | ||
| + | |||
}} | }} | ||
Revision as of 08:56, 14 June 2017
| |||||||||||
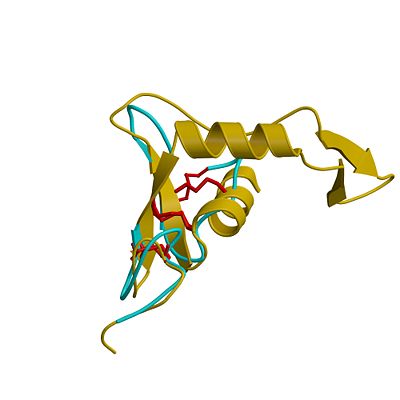
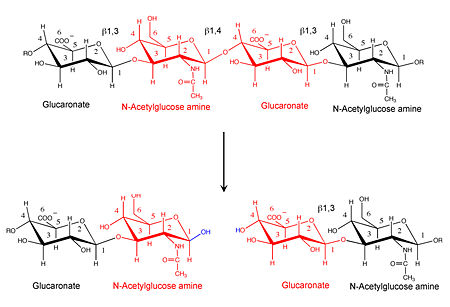
3D structures of hyaluronidase
Updated on 14-June-2017
Reference
- ↑ Chao KL, Muthukumar L, Herzberg O. Structure of human hyaluronidase-1, a hyaluronan hydrolyzing enzyme involved in tumor growth and angiogenesis. Biochemistry. 2007 Jun 12;46(23):6911-20. Epub 2007 May 16. PMID:17503783 doi:10.1021/bi700382g
- ↑ Ponnuraj K, Jedrzejas MJ. Mechanism of hyaluronan binding and degradation: structure of Streptococcus pneumoniae hyaluronate lyase in complex with hyaluronic acid disaccharide at 1.7 A resolution. J Mol Biol. 2000 Jun 16;299(4):885-95. PMID:10843845 doi:10.1006/jmbi.2000.3817
Created with the participation of Osnat Herzberg, Eran Hodis, Joel L. Sussman, Jaime Prilusky.