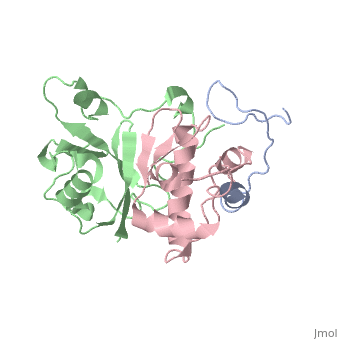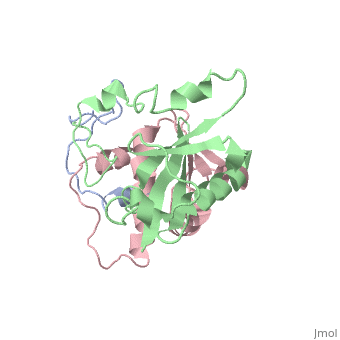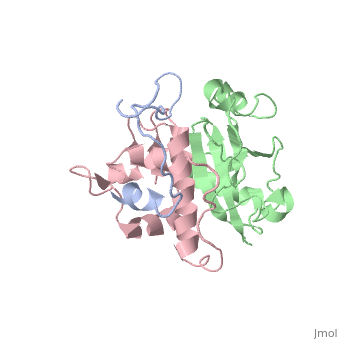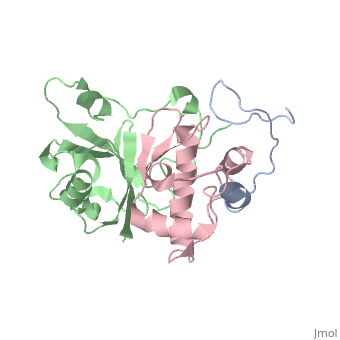
This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
HIV and accessory proteins
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | <StructureSection load='2MA9' size='350' side='right' scene='' caption=' | + | <StructureSection load='2MA9' size='350' side='right' scene='' caption=''> |
== Introduction == | == Introduction == | ||
Human immunodeficiency virus attacks the immune system by destroying CD4+ T cells, white blood cells that protect the body from infection. During HIV’s initial attack, it attaches to CD4 receptor cells injecting its RNA genetic material. The enzyme reverse transcriptase converts its’ RNA into DNA allowing HIV to use the CD4 cell’s machinery to replicate itself and travel through the body. As the virus attacks these cells, the immune system becomes weaker so the body is unable to fight infection, leading to the development of AIDS. Although HIV can be treated through the use of antiretroviral therapy there is currently no cure <ref name="Cycle">The HIV Life Cycle. (2015) Retrieved from [https://aidsinfo.nih.gov/education-materials/fact-sheets/19/73/the-hiv-life-cycle AIDSinfo]</ref>. HIV’s vast genetic variability makes treatment difficult. This variability is due to the high mutation and recombination rates of the reverse transcriptase enzyme causing HIV viral sequences to differ by up to 10% in each individual <ref name="Hemelaar">Hemelaar, J. (2012). The Origin and Diversity of the HIV-1 pandemic.In Trends in Molecular Medicine, 18(3):182-192 [http://doi.org/10.1016/j.molmed.2011.12.001 DOI:10.1016]</ref>. An estimated 36.9 million people were suffering from HIV around the world in 2014 <ref name="Global">Global HIV and AIDS Statistics. (2015). Retrieved from [http://www.avert.org/professionals/hiv-around-world/global-statistics Averting HIV and AIDS.]</ref>. | Human immunodeficiency virus attacks the immune system by destroying CD4+ T cells, white blood cells that protect the body from infection. During HIV’s initial attack, it attaches to CD4 receptor cells injecting its RNA genetic material. The enzyme reverse transcriptase converts its’ RNA into DNA allowing HIV to use the CD4 cell’s machinery to replicate itself and travel through the body. As the virus attacks these cells, the immune system becomes weaker so the body is unable to fight infection, leading to the development of AIDS. Although HIV can be treated through the use of antiretroviral therapy there is currently no cure <ref name="Cycle">The HIV Life Cycle. (2015) Retrieved from [https://aidsinfo.nih.gov/education-materials/fact-sheets/19/73/the-hiv-life-cycle AIDSinfo]</ref>. HIV’s vast genetic variability makes treatment difficult. This variability is due to the high mutation and recombination rates of the reverse transcriptase enzyme causing HIV viral sequences to differ by up to 10% in each individual <ref name="Hemelaar">Hemelaar, J. (2012). The Origin and Diversity of the HIV-1 pandemic.In Trends in Molecular Medicine, 18(3):182-192 [http://doi.org/10.1016/j.molmed.2011.12.001 DOI:10.1016]</ref>. An estimated 36.9 million people were suffering from HIV around the world in 2014 <ref name="Global">Global HIV and AIDS Statistics. (2015). Retrieved from [http://www.avert.org/professionals/hiv-around-world/global-statistics Averting HIV and AIDS.]</ref>. | ||
| Line 35: | Line 35: | ||
=== Viral protein R (Vpr) === | === Viral protein R (Vpr) === | ||
| - | < | + | <scene name='71/719207/Cv/2'>Figure 2. Viral protein R is involved with regulation, including movement of cDNA into the nucleus of the cell, promoting transcription, and stopping cell division</scene> (Protein Data Bank ID: [[1bde]]). |
| + | |||
| Line 62: | Line 63: | ||
=== Viral protein U (Vpu) === | === Viral protein U (Vpu) === | ||
| - | < | + | <scene name='71/719207/Cv/3'>Figure 3. Viral protein U is involved with the release of the virion</scene> (Protein Data Bank ID: [[1pi8]]). |
| Line 92: | Line 93: | ||
=== Negative regulatory factor (Nef) === | === Negative regulatory factor (Nef) === | ||
| - | < | + | <scene name='71/719207/Cv/4'>Figure 4. Negative regulatory factor (Nef) affects T cell regulation and downregulates host immune responses</scene> (Protein Data Bank: [[3rea]]). |
Negative regulatory factor (Nef) performs two functions in the host cell. The first is that it is involved in T cell activation. The second is that it maintains a persistent state of infection. | Negative regulatory factor (Nef) performs two functions in the host cell. The first is that it is involved in T cell activation. The second is that it maintains a persistent state of infection. | ||
| Line 122: | Line 123: | ||
=== Regulator of virion (Rev) === | === Regulator of virion (Rev) === | ||
| - | < | + | <scene name='71/719207/Cv/5'>Figure 5. Regulator of virion (Rev) exports unspliced RNA transcripts out of the nucleus</scene> (Protein Data Bank: [[4pmi]]). |
| + | |||
Regulator of virion (Rev) functions later in the pathway of HIV. It exports unspliced RNA out of the nucleus and into the cytoplasm. This unspliced RNA is thus ensured to be incorporated during assembly of the virion particle. | Regulator of virion (Rev) functions later in the pathway of HIV. It exports unspliced RNA out of the nucleus and into the cytoplasm. This unspliced RNA is thus ensured to be incorporated during assembly of the virion particle. | ||
| Line 147: | Line 149: | ||
=== Trans-activator of transcription (Tat) === | === Trans-activator of transcription (Tat) === | ||
| - | < | + | <scene name='71/719207/Cv/6'>Figure 6. Trans-activator of transcription (Tat) coupled with pTEFb, which stimulates transcription. Tat performs several other functions in the cell as well</scene> (Protein Data Bank ID: [[3mia]]). |
| + | |||
| Line 175: | Line 178: | ||
During a productive infection, HIV-1 is reactivated and the lytic cell cycle occurs. The host’s transcriptional and translational machinery is used. The proviral DNA is transcribed by the cellular RNA polymerase II from a single promoter in the 5’ LTR into a 9 kb viral RNA primary transcript. Trans-activator of transcription (Tat) and regulator of virion (Rev) both play a role in the regulation of viral gene expression. Tat interacts with TAR, which is located at the 5’ end of all HIV RNAs, and drastically increases the transcription rate from the HIV LTR <ref name="Shors" />. Rev binds to the Rev response element (RRE), which is a well-conserved, 350 nucleotide element that’s located in the env coding region of the viral genome <ref name="Fernandes">Fernandes, J., Jayaraman, B., & Frankel, A. (2012). The HIV-1 Rev response element: An RNA scaffold that directs the cooperative assembly of a homo-oligomeric ribonucleoprotein complex.[http://doi.org/10.4161/rna.9.1.18178 RNA Biology, 9(1), 6–11.]</ref>. This binding facilitates the export of unspliced and incompletely spliced viral RNAs from the nucleus to the cytoplasm. Once they are in the cytoplasm, some of the full-length viral messenger RNAs are packaged into virions, whereas others are spliced within the nucleus to form mRNAs that are translated by the host’s ribosome into different viral proteins. The gag gene and a combination of both the gag and pol genes together are translated into large precursor polyproteins that are cleaved by a virus encoded protease. The env protein, gp160, is transported through the trans Golgi apparatus where it is glycosylated and cleaved by a cellular protease into gp120 and gp41 to form mature envelope proteins that are targeted to the surface of the infected cell. The gp120 and gp41 are held together by covalent bonds. Tat and other smaller HIV proteins overlap with the structural genes but are in different open reading frames. Their mRNAs are made by alternative splicing of the structural gene mRNAs and are translated. The viral RNA, gag, pol, and env proteins, the virion components of HIV, are assembled at budding sites located at the cellular membrane. Two copies of the full viral genomes that contain the primer tRNA are packed into virus particles <ref name="Shors" />. | During a productive infection, HIV-1 is reactivated and the lytic cell cycle occurs. The host’s transcriptional and translational machinery is used. The proviral DNA is transcribed by the cellular RNA polymerase II from a single promoter in the 5’ LTR into a 9 kb viral RNA primary transcript. Trans-activator of transcription (Tat) and regulator of virion (Rev) both play a role in the regulation of viral gene expression. Tat interacts with TAR, which is located at the 5’ end of all HIV RNAs, and drastically increases the transcription rate from the HIV LTR <ref name="Shors" />. Rev binds to the Rev response element (RRE), which is a well-conserved, 350 nucleotide element that’s located in the env coding region of the viral genome <ref name="Fernandes">Fernandes, J., Jayaraman, B., & Frankel, A. (2012). The HIV-1 Rev response element: An RNA scaffold that directs the cooperative assembly of a homo-oligomeric ribonucleoprotein complex.[http://doi.org/10.4161/rna.9.1.18178 RNA Biology, 9(1), 6–11.]</ref>. This binding facilitates the export of unspliced and incompletely spliced viral RNAs from the nucleus to the cytoplasm. Once they are in the cytoplasm, some of the full-length viral messenger RNAs are packaged into virions, whereas others are spliced within the nucleus to form mRNAs that are translated by the host’s ribosome into different viral proteins. The gag gene and a combination of both the gag and pol genes together are translated into large precursor polyproteins that are cleaved by a virus encoded protease. The env protein, gp160, is transported through the trans Golgi apparatus where it is glycosylated and cleaved by a cellular protease into gp120 and gp41 to form mature envelope proteins that are targeted to the surface of the infected cell. The gp120 and gp41 are held together by covalent bonds. Tat and other smaller HIV proteins overlap with the structural genes but are in different open reading frames. Their mRNAs are made by alternative splicing of the structural gene mRNAs and are translated. The viral RNA, gag, pol, and env proteins, the virion components of HIV, are assembled at budding sites located at the cellular membrane. Two copies of the full viral genomes that contain the primer tRNA are packed into virus particles <ref name="Shors" />. | ||
The depletion of CD4+ helper T lymphocytes caused by HIV results in AIDS and a weakening of the immune system, permitting opportunistic infections to occur. The viral env proteins found on the surface of infected cells bind to uninfected cells causing the fusion of the plasma membrane, ultimately resulting in syncytia <ref name="Shors" />. | The depletion of CD4+ helper T lymphocytes caused by HIV results in AIDS and a weakening of the immune system, permitting opportunistic infections to occur. The viral env proteins found on the surface of infected cells bind to uninfected cells causing the fusion of the plasma membrane, ultimately resulting in syncytia <ref name="Shors" />. | ||
| - | + | </StructureSection> | |
== References == | == References == | ||
<references /> | <references /> | ||
Revision as of 09:16, 18 June 2017
| |||||||||||
References
- ↑ The HIV Life Cycle. (2015) Retrieved from AIDSinfo
- ↑ Hemelaar, J. (2012). The Origin and Diversity of the HIV-1 pandemic.In Trends in Molecular Medicine, 18(3):182-192 DOI:10.1016
- ↑ Global HIV and AIDS Statistics. (2015). Retrieved from Averting HIV and AIDS.
- ↑ 4.0 4.1 Origin of HIV & AIDS. (2015). Retrieved from Averting HIV and AIDS.
- ↑ Where Did HIV Come From? (2011). Retrieved from The AIDS Institute
- ↑ 6.0 6.1 Rose, Kristine M., Marin, Mariana, Kozak, Susan L., Kabat, David. (2004). The viral infectivity factor (Vif) of HIV-1 unveiled. TRENDS in Molecular Medicine, 10(6), 291-297.
- ↑ Morellet, N., Bouaziz, S., Petitjean, P., Roques, B. P. (2003). NMR Structure of the HIV-1 Regulatory Protein VPR. J. Mol. Biol, 327, 215-227.
- ↑ 8.0 8.1 Oie Solbak, Sara Marie, Reksten, Tove Ragna, Hahn, Friedrich, Wray, Victor, Henklein, Petra, Henklein, Peter, Halskau, Oyvind, Schubert, Ulrich, Fossen, Torgils. (2013). HIV-1 p6 - a structured to flexible multifunctional membrane-interacting protein. Biochimica et Biophysica Acta, 1828, 816-823.
- ↑ 9.0 9.1 Das, S.R., Jameel, S. (2005). Biology of the HIV Nef protein. Indian Journal of Medical Research, 121(4), 315-332.
- ↑ 10.0 10.1 Blissenbach, M., Grewe, B., Hoffman, B., Brandt, S., Uberla, K. (2010). Nuclear RNA export and packaging functions of HIV-1 Rev revisited. Journal of Virology, 84(13), 6598-604.
- ↑ 11.0 11.1 Das, A. T., Harwig, A., & Berkhout, B. (2011). The HIV-1 Tat Protein Has a Versatile Role in Activating Viral Transcription. Journal of Virology, 85(18), 9506–9516.
- ↑ Yao, S., Torres, A.M., Azad, A.A., Macreadie, I.G., Norton, R.S. (1998) Solution structure of peptides from HIV-1 VPR protein that cause membrane permeabilization and growth arrest. J.Pebt.Sci. 4.426-435
- ↑ Demirov, Dimiter G., Orenstein, Jan M., and Freed, Eric O., (2002). The late domain of Human Immunodeficiency Virus type 1 promotes virus release in a cell type-dependent manner. Journal of Virology. 71(1), 105-117.
- ↑ 14.0 14.1 14.2 14.3 Shors, Teri. (2011). Understanding Viruses. (2nd ed., pp. 507-511). Oshkosh, Wisconsin: Jones and Bartlett.
- ↑ Fernandes, J., Jayaraman, B., & Frankel, A. (2012). The HIV-1 Rev response element: An RNA scaffold that directs the cooperative assembly of a homo-oligomeric ribonucleoprotein complex.RNA Biology, 9(1), 6–11.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Erin Bolger, Alexander Berchansky, Joel L. Sussman