Lipase
From Proteopedia
(Difference between revisions)
| Line 72: | Line 72: | ||
**[[1hpl]] – hLip – horse <br /> | **[[1hpl]] – hLip – horse <br /> | ||
**[[1hlg]] – hLip – human - gastric<br /> | **[[1hlg]] – hLip – human - gastric<br /> | ||
| - | **[[3jw8]], [[3hju]] – mono-glyceride hLip<br /> | ||
**[[1jmy]] – hBSSL <br /> | **[[1jmy]] – hBSSL <br /> | ||
**[[1akn]] – cBSSL – cattle <br /> | **[[1akn]] – cBSSL – cattle <br /> | ||
| Line 81: | Line 80: | ||
**[[1gpl]] – Lip – Guinea pig<br /> | **[[1gpl]] – Lip – Guinea pig<br /> | ||
**[[3zpx]] – Lip – ''Ustilago maydis''<br /> | **[[3zpx]] – Lip – ''Ustilago maydis''<br /> | ||
| + | **[[4l3w]] – Lip catalytic domain – bread mold<br /> | ||
| + | **[[4zrd]], [[4zre]] – Lip1 (mutant) – dandruff fungus<br /> | ||
*Prokaryote lipase: | *Prokaryote lipase: | ||
| Line 86: | Line 87: | ||
**[[3guu]], [[1lbs]], [[1lbt]], [[1tca]], [[1tcb]], [[1tcc]] – CaLipA – ''Candida antarctica''<br /> | **[[3guu]], [[1lbs]], [[1lbt]], [[1tca]], [[1tcb]], [[1tcc]] – CaLipA – ''Candida antarctica''<br /> | ||
**[[2veo]] – CaLipA – closed state<br /> | **[[2veo]] – CaLipA – closed state<br /> | ||
| - | **[[4k6g]] – CaLipB<br /> | + | **[[4k6g]], [[4zv7]], [[3w9b]], [[5a6v]], [[5a71]] – CaLipB<br /> |
**[[3icv]], [[4k5q]], [[4k6h]], [[4k6k]] – CaLipB (mutant) <br /> | **[[3icv]], [[4k5q]], [[4k6h]], [[4k6k]] – CaLipB (mutant) <br /> | ||
**[[1gz7]], [[1lpm]], [[1lps]]– CrLip 2 – ''Candida rugosa'' - closed state<br /> | **[[1gz7]], [[1lpm]], [[1lps]]– CrLip 2 – ''Candida rugosa'' - closed state<br /> | ||
| Line 97: | Line 98: | ||
**[[2fx5]] – Lip – ''Pseudomonas mendocina''<br /> | **[[2fx5]] – Lip – ''Pseudomonas mendocina''<br /> | ||
**[[1yzf]] – Lip – ''Enterococcus faecalis''<br /> | **[[1yzf]] – Lip – ''Enterococcus faecalis''<br /> | ||
| - | **[[1dt3]], [[1dt5]], [[1dte]], [[1du4]], [[1ein]], [[1tib]], [[4dyh]], [[4ea6]], [[4flf]], [[4gbg]], [[4gwl]] - TlLip - ''Thermomyces lanuginose''<br /> | + | **[[1dt3]], [[1dt5]], [[1dte]], [[1du4]], [[1ein]], [[1tib]], [[4dyh]], [[4ea6]], [[4flf]], [[4gbg]], [[4gwl]], [[4zgb]] - TlLip - ''Thermomyces lanuginose''<br /> |
| + | **[[5ap9]] – TlLip (mutant)<br /> | ||
**[[1jfr]] – Lip – ''Streptomyces exfoliates''<br /> | **[[1jfr]] – Lip – ''Streptomyces exfoliates''<br /> | ||
| + | **[[5mal]] – Lip – ''Streptomyces rimosus''<br /> | ||
**[[1oil]] – BcLip - ''Burkholderia cepacia''<br /> | **[[1oil]] – BcLip - ''Burkholderia cepacia''<br /> | ||
**[[2lip]] – BcLip – open state<br /> | **[[2lip]] – BcLip – open state<br /> | ||
| Line 112: | Line 115: | ||
**[[3lip]], [[3a6z]] - Lip - ''Pseudomonas cepacia'' – open state<br /> | **[[3lip]], [[3a6z]] - Lip - ''Pseudomonas cepacia'' – open state<br /> | ||
**[[1qge]], [[1tah]] – Lip – ''Pseudomonas glumae''<br /> | **[[1qge]], [[1tah]] – Lip – ''Pseudomonas glumae''<br /> | ||
| - | **[[2w22]] – | + | **[[2w22]] – GtLip – ''Geobacillus thermocatenulatus''<br /> |
| - | **[[1ji3]], [[1ku0]], [[4fmp]] – | + | **[[5ce5]] – GtLip (mutant)<br /> |
| + | **[[1ji3]], [[1ku0]], [[4fmp]], [[4x6u]]– BstLip – ''Bacillus stearothermophilus''<br /> | ||
| + | **[[4x71]], [[4x7b]], [[4x85]] – BstLip (mutant)<br /> | ||
**[[1ah7]] - Lip – ''Bacillus cereus''<br /> | **[[1ah7]] - Lip – ''Bacillus cereus''<br /> | ||
| - | **[[2qxt]], [[2qxu]], [[1isp]], [[1i6w]], [[4fdm]] - BsLip – ''Bacillus subtilis''<br /> | + | **[[2qxt]], [[2qxu]], [[1isp]], [[1i6w]], [[4fdm]], [[5ct5]], [[5ct6]] - BsLip – ''Bacillus subtilis''<br /> |
| - | **[[3d2a]], [[3d2b]], [[3d2c]], [[1t2n]], [[1t4m]], [[3qmm]], [[3qzu]], [[4fkb]] - BsLip (mutant) <br /> | + | **[[3d2a]], [[3d2b]], [[3d2c]], [[1t2n]], [[1t4m]], [[3qmm]], [[3qzu]], [[4fkb]], [[5ct8]] - BsLip (mutant) <br /> |
**[[2ory]] – Lip – ''Photobacterium lypoliticum''<br /> | **[[2ory]] – Lip – ''Photobacterium lypoliticum''<br /> | ||
**[[2z5g]], [[2dsn]] – GzLip T1 – ''Geobacillus zalihae''<br /> | **[[2z5g]], [[2dsn]] – GzLip T1 – ''Geobacillus zalihae''<br /> | ||
| Line 125: | Line 130: | ||
**[[3uue]], [[3uuf]] – Lip – ''Malassezia globosa''<br /> | **[[3uue]], [[3uuf]] – Lip – ''Malassezia globosa''<br /> | ||
**[[4hs9]] – Lip – ''Proteus mirabilis''<br /> | **[[4hs9]] – Lip – ''Proteus mirabilis''<br /> | ||
| + | **[[4opm]] – Lip – ''Acinetobacter baumannii''<br /> | ||
| + | **[[5ah0]] – Lip – ''Pelosinus fermentans''<br /> | ||
| + | **[[5ah1]] – Lip – ''Clostridium botulinum''<br /> | ||
| + | **[[5ch8]] – Lip (mutant) – ''Penicillium cyclopium''<br /> | ||
*Lipase/colipase complexes. The colipase is a co-enzyme whose binding to lipase optimizes the enzymatic activity | *Lipase/colipase complexes. The colipase is a co-enzyme whose binding to lipase optimizes the enzymatic activity | ||
| Line 150: | Line 159: | ||
*Lipase + inhibitors | *Lipase + inhibitors | ||
| - | **[[3jwe]], [[3pe6]] - mono-glyceride hLip + SAR629 – covalent inhibitor<br /> | ||
**[[3l1h]] – EstE5(LIPE)+FeCl3 – noninvasive inhibitor<br /> | **[[3l1h]] – EstE5(LIPE)+FeCl3 – noninvasive inhibitor<br /> | ||
**[[3l1i]], [[3l1j]] - EstE5(LIPE)+CuSO4 – noninvasive inhibitor<br /> | **[[3l1i]], [[3l1j]] - EstE5(LIPE)+CuSO4 – noninvasive inhibitor<br /> | ||
| Line 173: | Line 181: | ||
**[[4kjx]] - TlLip + nitrobenzaldehyde + lauric acid<br /> | **[[4kjx]] - TlLip + nitrobenzaldehyde + lauric acid<br /> | ||
**[[4n8s]] - TlLip + nitrobenzaldehyde + ethylacetoacetate<br /> | **[[4n8s]] - TlLip + nitrobenzaldehyde + ethylacetoacetate<br /> | ||
| + | **[[4s0x]] – TlLip + lauric acid<br /> | ||
*Lipase conjugated with analogs to its reaction intermediates | *Lipase conjugated with analogs to its reaction intermediates | ||
| Line 191: | Line 200: | ||
**[[4ke7]], [[4ke8]], [[4ke9]] – BaMAGL + ligand<br /> | **[[4ke7]], [[4ke8]], [[4ke9]] – BaMAGL + ligand<br /> | ||
**[[4ke6]], [[4kea]] – BaMAGL (mutant) + ligand<br /> | **[[4ke6]], [[4kea]] – BaMAGL (mutant) + ligand<br /> | ||
| - | **[[3jwe]], [[3pe6]] – hMAGL + inhibitor<br /> | + | **[[3jwe]], [[3pe6]], [[4uuq]] – hMAGL + inhibitor<br /> |
| + | **[[4zwn]] – yMAGL – yeast<br /> | ||
| + | **[[4zxf]] – yMAGLip + substrate analog<br /> | ||
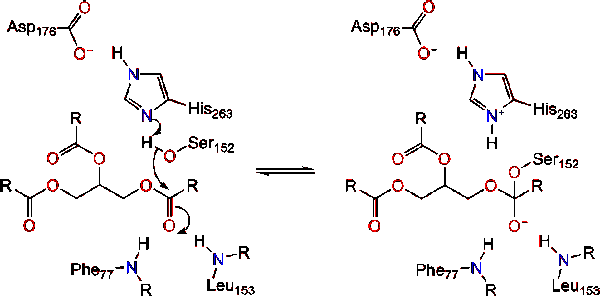
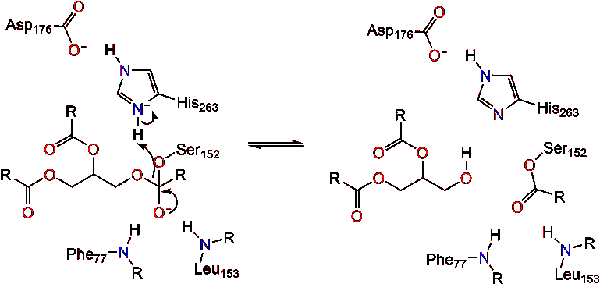
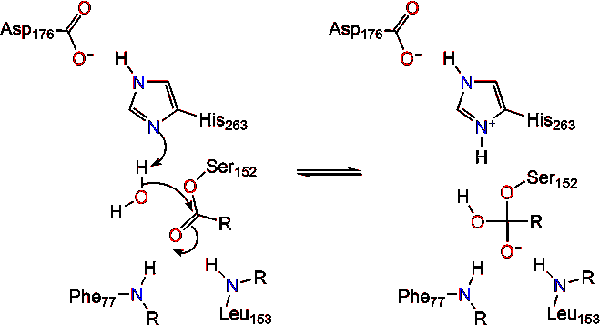
*Lipase with substrate bound at active site | *Lipase with substrate bound at active site | ||
Revision as of 08:31, 4 July 2017
| |||||||||||
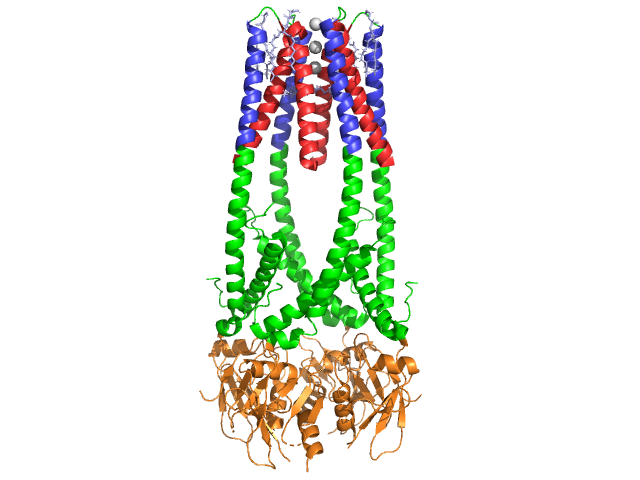
3D Structures of Lipase
Updated on 04-July-2017
References
- ↑ [1] 1HPL PDB SUM
- ↑ [2] A cross-linked complex between horse pancreatic lipase and colipase
- ↑ [3] History of Lipids
- ↑ [4] 1HPL PDB
- ↑ http://www.pdb.org/pdb/explore/explore.do?structureId=1HPL
- ↑ http://www.pdb.org/pdb/explore/remediatedSequence.do?structureId=1HPL
- ↑ http://www.springerlink.com/content/g5h1613440115701/fulltext.pdf
- ↑ Fundamentals of Biochemistry...
- ↑ Thomas, A. etc. "Role of the Lid Hydrophobicity Pattern in Pancreatic Lipase Activity", The Journal of Biological Chemistry, 2005 September 22; 270 (48): 40074-40083.
- ↑ "Colipase". Wikipedia: The Free Encyclopedia. 5 July 2011 [5]
- ↑ "Colipase Residues..."
- ↑ Fundamentals of Biochemistry...
- ↑ Crandall,W., Lowe, M. "Colipase Residues Glu64 and Arg65 Are Essential for Normal Lipase-mediated Fat Digestion in the Presence of Bile Salt Micelles" Journal of Biological Chemistry, 2001, (276) 12505-12512
- ↑ van Tilbeurgh H, etc."Structure of the pancreatic lipase-procolipase complex", 1992 Sep 10;359(6391):159-62. PMID:1522902.[6]
- ↑ http://www.pdb.org/pdb/explore/explore.do?structureId=1ETH
- ↑ http://www.nature.com/nature/journal/v362/n6423/abs/362814a0.html
- ↑ Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science. 1991 Aug 23;253(5022):872-9. PMID:1678899
- ↑ Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, et al.. The alpha/beta hydrolase fold. Protein Eng. 1992 Apr;5(3):197-211. PMID:1409539
- ↑ Bourne Y, Martinez C, Kerfelec B, Lombardo D, Chapus C, Cambillau C. Horse pancreatic lipase. The crystal structure refined at 2.3 A resolution. J Mol Biol. 1994 May 20;238(5):709-32. PMID:8182745 doi:http://dx.doi.org/10.1006/jmbi.1994.1331
- ↑ [7] 1LPB PDB SUM
- ↑ "Pancreatic lipase". Wikipedia: The Free Encyclopedia. 7 Nov 2011 [8]
- ↑ Kordik, C., Reitz, A. "Pharmacological Treatment of Obesity: Therapeutic Strategies" Journal of Medicinal Chemistry, 1999 (42).
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Quinn R. Murray, Natalie Ziegler, Stephanie Schell, David Canner, Alexander Berchansky, Katelyn Clark, Eric Martz, Leben Tadesse, Joel L. Sussman, Eran Hodis