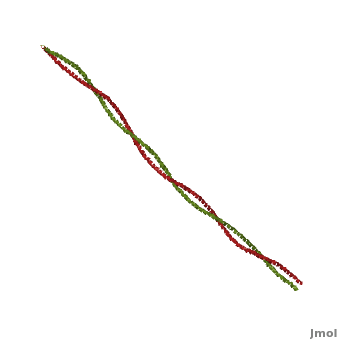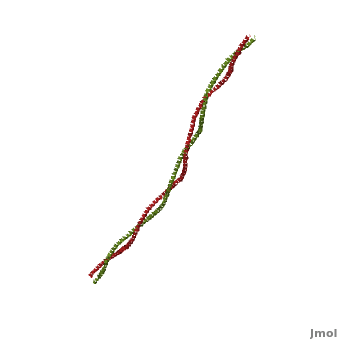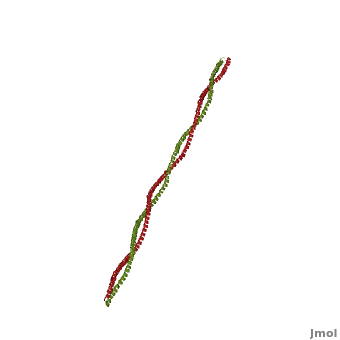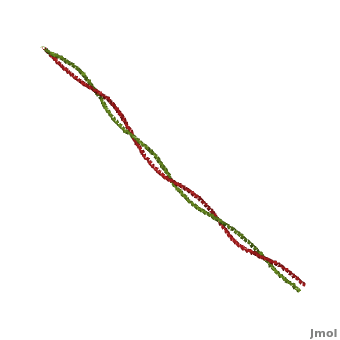
Tropomyosin
From Proteopedia
(Difference between revisions)
| Line 31: | Line 31: | ||
=== Post-Translational Modifications === | === Post-Translational Modifications === | ||
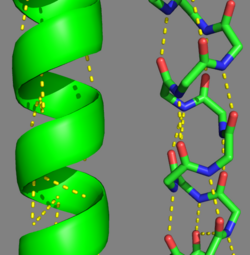
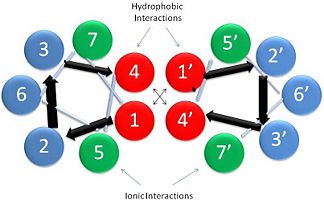
| - | There are two types of post-translational modifications to tropomyosin: phosphorylation acetylation<ref name="Gunning"/>. Phosphorylation occurs on amino acid <scene name=' | + | There are two types of post-translational modifications to tropomyosin: phosphorylation acetylation<ref name="Gunning"/>. Phosphorylation occurs on amino acid <scene name='41/410306/Tropomyosin_dimer_ser229/1'>Ser-229</scene><ref name="Gunning"/>. This phosphorylation is occurs as a result of oxidative stress, which is associated with actin remodeling and recruitment of additional tropomyosin into stress fibers<ref name="Gunning"/>. The acetylation occurs on the N-terminus of the N-terminal methionine, which is essential for: coiled-coil stability, overlap formation and actin binding<ref name="Frye"/><ref name="Gunning"/>. |
=== Evolutionary Conservation === | === Evolutionary Conservation === | ||
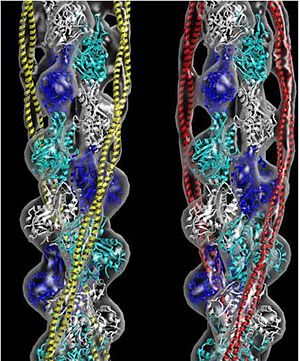
Tropomyosin is highly conserved actin binding protein, which is found in Eukarya from the animal kingdom to yeast, with the exception of plants<ref name="Gunning"/>. The earliest characterization of tropomyosin gene lineage was in yeast (budding and fission yeast)<ref name="Gunning"/><ref name="Drees">PMID:7844152</ref>. These genes are known as [http://en.wikipedia.org/wiki/TPM1 TPM1] and [http://en.wikipedia.org/wiki/TPM2 TPM2], respectively, and share 64.5% sequence identity<ref name="Gunning"/><ref name="Drees"/>. As we move away from unicellular organisms and into multicellular invertebrates, tropomyosin genes in nematodes slightly diverge from yeast, but have 85-90% sequence identity between their genes<ref name="Gunning"/>. Further analysis for vertebrates show there are four genes that generate over 40 known mammalian isoforms of tropomyosin, which are synthesized by exon splicing<ref name="Gunning"/>. The slight evolution change of tropomyosin has occurred as a result of the increasing need of tropomyosin to function in different systems, but tropomyosin has evolutionarily stayed well conserved because of the basic structural pressures imposed on the protein<ref name="Gunning"/>. It is interesting to note, the region with the least conservation has been in the N and C terminus. This is the result of head-tail interactions changing to accommodate different polymer confirmations along actin for different functions<ref name="Gunning"/>. (To see this evolutionary conservation, go to the top right image of the web page and click on the "show" link, which is to the right of "Evolutionary Conservation".) | Tropomyosin is highly conserved actin binding protein, which is found in Eukarya from the animal kingdom to yeast, with the exception of plants<ref name="Gunning"/>. The earliest characterization of tropomyosin gene lineage was in yeast (budding and fission yeast)<ref name="Gunning"/><ref name="Drees">PMID:7844152</ref>. These genes are known as [http://en.wikipedia.org/wiki/TPM1 TPM1] and [http://en.wikipedia.org/wiki/TPM2 TPM2], respectively, and share 64.5% sequence identity<ref name="Gunning"/><ref name="Drees"/>. As we move away from unicellular organisms and into multicellular invertebrates, tropomyosin genes in nematodes slightly diverge from yeast, but have 85-90% sequence identity between their genes<ref name="Gunning"/>. Further analysis for vertebrates show there are four genes that generate over 40 known mammalian isoforms of tropomyosin, which are synthesized by exon splicing<ref name="Gunning"/>. The slight evolution change of tropomyosin has occurred as a result of the increasing need of tropomyosin to function in different systems, but tropomyosin has evolutionarily stayed well conserved because of the basic structural pressures imposed on the protein<ref name="Gunning"/>. It is interesting to note, the region with the least conservation has been in the N and C terminus. This is the result of head-tail interactions changing to accommodate different polymer confirmations along actin for different functions<ref name="Gunning"/>. (To see this evolutionary conservation, go to the top right image of the web page and click on the "show" link, which is to the right of "Evolutionary Conservation".) | ||
Revision as of 08:30, 9 July 2017
| |||||||||||
3D structures of Tropomyosin
Updated on 09-July-2017
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 1.19 1.20 1.21 1.22 1.23 1.24 Tropomyosins. I. Gunning, Peter, 1950- II. Series.[DNLM: 1. Tropomyosin. W1 AD559 v.644 2008 / WE 500 T856 2008]
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Frye J, Klenchin VA, Rayment I. Structure of the tropomyosin overlap complex from chicken smooth muscle: insight into the diversity of N-terminal recognition . Biochemistry. 2010 Jun 15;49(23):4908-20. PMID:20465283 doi:10.1021/bi100349a
- ↑ 3.0 3.1 3.2 Whitby FG, Phillips GN Jr. Crystal structure of tropomyosin at 7 Angstroms resolution. Proteins. 2000 Jan 1;38(1):49-59. PMID:10651038
- ↑ 4.0 4.1 4.2 Clayton JE, Sammons MR, Stark BC, Hodges AR, Lord M. Differential regulation of unconventional fission yeast myosins via the actin track. Curr Biol. 2010 Aug 24;20(16):1423-31. Epub 2010 Aug 12. PMID:20705471 doi:10.1016/j.cub.2010.07.026
- ↑ 5.0 5.1 Stark BC, Sladewski TE, Pollard LW, Lord M. Tropomyosin and myosin-II cellular levels promote actomyosin ring assembly in fission yeast. Mol Biol Cell. 2010 Mar 15;21(6):989-1000. Epub 2010 Jan 28. PMID:20110347 doi:10.1091/mbc.E09-10-0852
- ↑ 6.0 6.1 Drees B, Brown C, Barrell BG, Bretscher A. Tropomyosin is essential in yeast, yet the TPM1 and TPM2 products perform distinct functions. J Cell Biol. 1995 Feb;128(3):383-92. PMID:7844152
- ↑ 7.0 7.1 7.2 7.3 7.4 Lehman W, Galinska-Rakoczy A, Hatch V, Tobacman LS, Craig R. Structural basis for the activation of muscle contraction by troponin and tropomyosin. J Mol Biol. 2009 May 15;388(4):673-81. Epub 2009 Mar 31. PMID:19341744 doi:10.1016/j.jmb.2009.03.060
- ↑ 8.0 8.1 Tyska MJ, Warshaw DM. The myosin power stroke. Cell Motil Cytoskeleton. 2002 Jan;51(1):1-15. PMID:11810692 doi:10.1002/cm.10014
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Gregory Hoeprich, Jaime Prilusky, David Canner, Joel L. Sussman