This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Polygalacturonase
From Proteopedia
| Line 9: | Line 9: | ||
== Structural highlights == | == Structural highlights == | ||
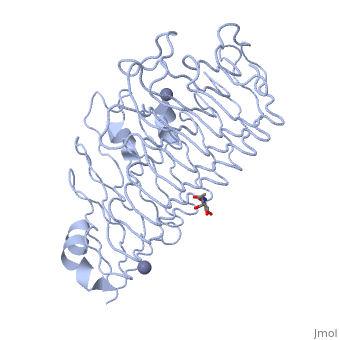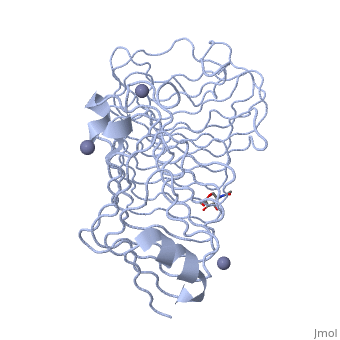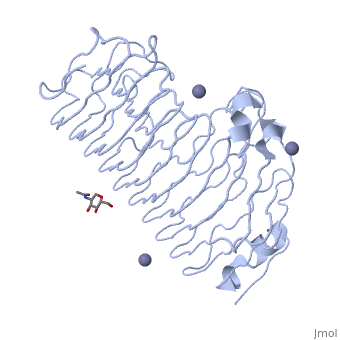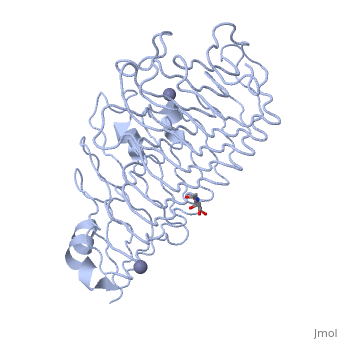
| - | + | The secondary structure of PGs is comprised of a ten turn right handed beta helix domain along with two loop regions that together form the substrate-binding cleft, which appears to have a tunnel-like shape. The active site of PGs is found between the two looped regions of the protein. Located within the looped regions are two conserved aspartate residues that are predicted to participate in catalytic activity. | |
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 18:36, 12 July 2017
|
Contents |
Function
PGs cleave α-(1-4) – glycosidic bonds between consecutive galacturonic acid residues.
Disease
Relevance
Structural highlights
The secondary structure of PGs is comprised of a ten turn right handed beta helix domain along with two loop regions that together form the substrate-binding cleft, which appears to have a tunnel-like shape. The active site of PGs is found between the two looped regions of the protein. Located within the looped regions are two conserved aspartate residues that are predicted to participate in catalytic activity.
</StructureSection>
References
Proteopedia Page Contributors and Editors (what is this?)
Joel L. Sussman, Krishna Amin, Michal Harel, Marilyn Yoder, OCA, Jaime Prilusky