Urate Oxidase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
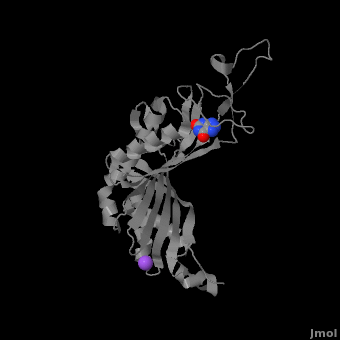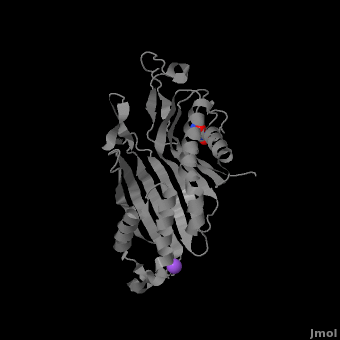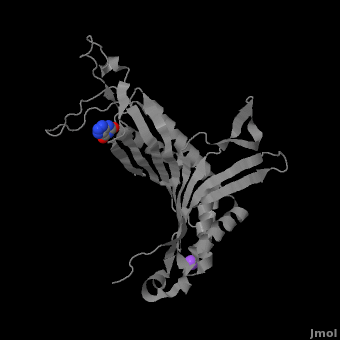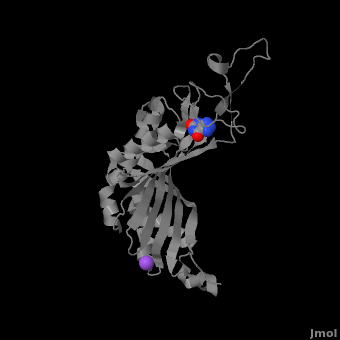
| - | <StructureSection load='3bk8' size=' | + | <StructureSection load='3bk8' size='400' side='right' caption='Monomer of the tetrameric urate oxidase complex with 8-azaxanthine and Na+ ion (purple) [[3bk8]]' scene='Sandbox_185/Urate_oxidase_basic_monomer/1' > |
== Introduction == | == Introduction == | ||
| Line 9: | Line 9: | ||
Within the active site, two residues, <scene name='Sandbox_185/Urate_oxidase_basic_monomer/3'>Arg 176</scene> and <scene name='Sandbox_185/Urate_oxidase_basic_monomer/2'>Gln 228</scene>, are responsible for hydrogen-binding the substrate (uric acid).<ref name="gabison"/> Approximately 0.33 nm above the ligand, Asn 254 and Thr 57* (* indicates a residue from a different subunit), hydrogen-bond the molecular oxygen and the catalytic water molecule, depending on the step of the reaction.<ref name="gabison"/> The catalytic water molecule and the γ oxygen of Thr 57* are the beginning of a proton transfer chain that also involves Lys 10*, His 256 and two other water molceules, ending at N9 of uric acid.<ref name="gabison"/> | Within the active site, two residues, <scene name='Sandbox_185/Urate_oxidase_basic_monomer/3'>Arg 176</scene> and <scene name='Sandbox_185/Urate_oxidase_basic_monomer/2'>Gln 228</scene>, are responsible for hydrogen-binding the substrate (uric acid).<ref name="gabison"/> Approximately 0.33 nm above the ligand, Asn 254 and Thr 57* (* indicates a residue from a different subunit), hydrogen-bond the molecular oxygen and the catalytic water molecule, depending on the step of the reaction.<ref name="gabison"/> The catalytic water molecule and the γ oxygen of Thr 57* are the beginning of a proton transfer chain that also involves Lys 10*, His 256 and two other water molceules, ending at N9 of uric acid.<ref name="gabison"/> | ||
| - | + | *<scene name='38/382957/Cv/3'>Whole active site</scene>. | |
== Function == | == Function == | ||
Revision as of 10:00, 24 July 2017
| |||||||||||
3D structures of urate oxidase
Updated on 24-July-2017
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 Gabison L, Prange T, Colloc'h N, El Hajji M, Castro B, Chiadmi M. Structural analysis of urate oxidase in complex with its natural substrate inhibited by cyanide: mechanistic implications. BMC Struct Biol. 2008 Jul 20;8:32. PMID:18638417 doi:10.1186/1472-6807-8-32
- ↑ 2.0 2.1 2.2 2.3 Colloc'h N, el Hajji M, Bachet B, L'Hermite G, Schiltz M, Prange T, Castro B, Mornon JP. Crystal structure of the protein drug urate oxidase-inhibitor complex at 2.05 A resolution. Nat Struct Biol. 1997 Nov;4(11):947-52. PMID:9360612
- ↑ 3.0 3.1 Colloc'h N, Girard E, Dhaussy a, Kahn R, Ascone I, Mezouar M, Fourme R. High pressure macromolecular crystallography: the 140-MPa resolution of urate oxidase, a 135-kDa tetrameric assembly. Biochemica et Biophysica Acta - Proteins and Proteomics. 2006 March;1764:3.
- ↑ 4.0 4.1 Wu XW, Muzny DM, Lee CC, Caskey CT. Two independent mutational events in the loss of urate oxidase during hominoid evolution. J Mol Evol. 1992 Jan;34(1):78-84. PMID:1556746
- ↑ 5.0 5.1 5.2 5.3 Renyi I, Bardi E, Udvardi E, Kovacs G, Bartyik K, Kajtar P, Masat P, Nagy K, Galantai I, Kiss C. Prevention and treatment of hyperuricemia with rasburicase in children with leukemia and non-Hodgkin's lymphoma. Pathol Oncol Res. 2007;13(1):57-62. Epub 2007 Mar 27. PMID:17387390 doi:PAOR.2007.13.1.0057
Proteopedia Page Contributors and Editors (what is this?)
Sonja Senekovic, Michal Harel, Alexander Berchansky, David Canner, Andrea Gorrell, Joel L. Sussman