Sandbox Reserved 1053
From Proteopedia
| Line 9: | Line 9: | ||
<StructureSection load='2KJB' size='340' frame='true' side='right' caption='The dimer CzrA' scene=''> | <StructureSection load='2KJB' size='340' frame='true' side='right' caption='The dimer CzrA' scene=''> | ||
| - | + | ==Czr Operon== | |
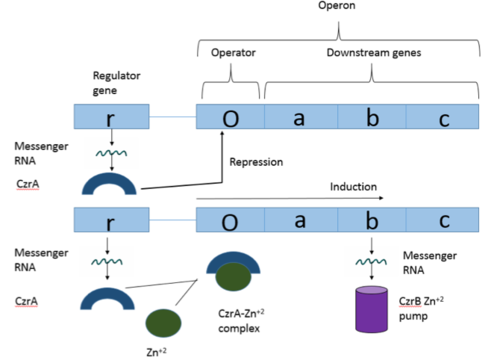
The Chromosome Determined Zinc Responsible (Czr) operon acts as described above, with Czr A acting as a regulator protein. As such, Czr A is responsible for controlling the transcription of the rest of the operon and by extension, the transport of Zn <sup>2+</sup> out of the cell; the role of Czr A in the Czr operon is described in further detail as part of the explanation of biological function. In addition to being a component of an operon, Czr A is also considered to be a metal sensor protein. While the immediate function of Czr A is gene regulation, this serves the larger purpose of acting to maintain an appropriate concentration of Zn <sup>2+</sup> in the cell. | The Chromosome Determined Zinc Responsible (Czr) operon acts as described above, with Czr A acting as a regulator protein. As such, Czr A is responsible for controlling the transcription of the rest of the operon and by extension, the transport of Zn <sup>2+</sup> out of the cell; the role of Czr A in the Czr operon is described in further detail as part of the explanation of biological function. In addition to being a component of an operon, Czr A is also considered to be a metal sensor protein. While the immediate function of Czr A is gene regulation, this serves the larger purpose of acting to maintain an appropriate concentration of Zn <sup>2+</sup> in the cell. | ||
| Line 18: | Line 18: | ||
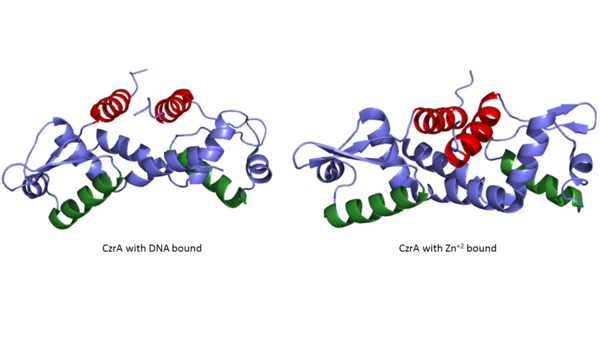
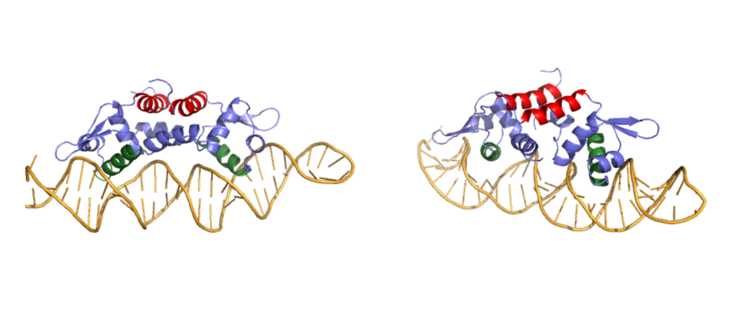
18177-18182.</ref>. The Czr operon contains genes for the proteins Czr A and [http://proteopedia.org/wiki/index.php/3byr Czr B]. Czr B is a Zinc transport protein which moves Zn<sup>2+</sup> out of a cell while Czr A regulates this process by controlling expression level of Czr B. When relatively low amounts of zinc are present in the cell Czr A will bind to DNA, preventing the progression of RNA polymerase and thus inhibiting expression of Czr B. Decreased expression of Czr B results in the ability of the cell to retain Zn<sup>2+</sup> more readily. Because Czr A and Czr B are transcribed as part of the same operon, an inhibitor of Czr A must be readily available to allow full transcription of Czr B when necessary. Czr A is noncompetitively inhibited by the binding of two Zn<sup>2+</sup> ions, which is ideal in that this allows expression of Czr B, a Zn<sup>2+</sup> transporter to be dependent on the relative amount of Zn<sup>2+</sup> in the cell. Czr A displays two different conformations; the first binds DNA and has relatively low affinity for Zn<sup>2+</sup> (PDB code: 2kjb). In this conformation the <scene name='69/694220/A5_helices__dna_binding/1'>a5 helices are aligned</scene>. Binding of zinc drives a conformational change (PDB code: 2kjc) in which the <scene name='69/694220/A5_helices_dna_binding/1'>a5 helices become unaligned</scene>, lowering the affinity for DNA. | 18177-18182.</ref>. The Czr operon contains genes for the proteins Czr A and [http://proteopedia.org/wiki/index.php/3byr Czr B]. Czr B is a Zinc transport protein which moves Zn<sup>2+</sup> out of a cell while Czr A regulates this process by controlling expression level of Czr B. When relatively low amounts of zinc are present in the cell Czr A will bind to DNA, preventing the progression of RNA polymerase and thus inhibiting expression of Czr B. Decreased expression of Czr B results in the ability of the cell to retain Zn<sup>2+</sup> more readily. Because Czr A and Czr B are transcribed as part of the same operon, an inhibitor of Czr A must be readily available to allow full transcription of Czr B when necessary. Czr A is noncompetitively inhibited by the binding of two Zn<sup>2+</sup> ions, which is ideal in that this allows expression of Czr B, a Zn<sup>2+</sup> transporter to be dependent on the relative amount of Zn<sup>2+</sup> in the cell. Czr A displays two different conformations; the first binds DNA and has relatively low affinity for Zn<sup>2+</sup> (PDB code: 2kjb). In this conformation the <scene name='69/694220/A5_helices__dna_binding/1'>a5 helices are aligned</scene>. Binding of zinc drives a conformational change (PDB code: 2kjc) in which the <scene name='69/694220/A5_helices_dna_binding/1'>a5 helices become unaligned</scene>, lowering the affinity for DNA. | ||
| - | + | == Zinc Binding == | |
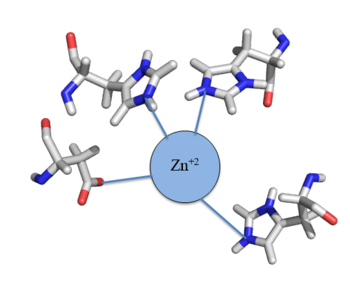
Zinc acts as an inhibitor to Czr A<ref name="critical"/>, thus preventing transcriptional repression of Czr B and allowing Zn<sup>2+</sup> transport out of the cell. This allows for zinc transport to essentially be self regulated. That is, when zinc concentration in the cell is relatively high, zinc ions bind to Czr A, causing a conformational change which releases the bound DNA. DNA without Czr A bound is free to be transcribed and Czr B is again expressed, allowing for Zn<sup>2+</sup> transport out of the cell. At low Zn<sup>2+</sup> concentrations, CzrA represses RNA Polymerase activity, and Zn<sup>2+</sup> ions are maintained inside the cell. | Zinc acts as an inhibitor to Czr A<ref name="critical"/>, thus preventing transcriptional repression of Czr B and allowing Zn<sup>2+</sup> transport out of the cell. This allows for zinc transport to essentially be self regulated. That is, when zinc concentration in the cell is relatively high, zinc ions bind to Czr A, causing a conformational change which releases the bound DNA. DNA without Czr A bound is free to be transcribed and Czr B is again expressed, allowing for Zn<sup>2+</sup> transport out of the cell. At low Zn<sup>2+</sup> concentrations, CzrA represses RNA Polymerase activity, and Zn<sup>2+</sup> ions are maintained inside the cell. | ||
Revision as of 16:51, 9 August 2017
CzrA: A Zinc Dependent Transcriptional Regulator
|
Background
Operon Overview
Operons are a critical genetic component of most prokaryotic cells. There are many different operons, responsible for the production of proteins with a wide range of functions. The most well-known and studied operons are the Lac and Trp operons, responsible for producing enzymes which metabolize lactose and tryptophan respectively. Despite many differences in each operon and the proteins that they encode, operons all function in the same general manner. Structurally, each operon contains a regulator, an operator, and one or more structural genes. The regulator gene codes for a protein responsible for managing the expression level of the structural genes. The operator contains the binding sequence for RNA polymerase and is the site where transcription begins. Lastly, the structural genes code for proteins to be used elsewhere. The regulator protein (produced as a result of expression of the regulator gene) most often acts in a repressive manner, though this is not always the case. That is, the regulator protein will bind to the operator, inhibiting the binding and/or progression of RNA polymerase to the structural genes, thus inhibiting transcription of the genes into mRNA. If the regulator protein were always active, there could never be adequate expression of the structural genes, so there must be a way to inactive the regulator protein, thus enabling expression of the structural genes. This is usually achieved through the binding of an inhibitor to the regulator protein. Since regulator proteins are DNA binding proteins, often this inhibition is allosteric rather than competitive, that is the inhibitor is not something that mimics DNA and binds to the active site physically blocking DNA from binding. Rather, the inhibitor of the regulator binds to somewhere other than the active site of the protein, changing it in some way which decreases the proteins affinity or ability to bind DNA and repress transcription.
Structure Testing Area
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Arunkumar A., Campanello G., Giedroc D. (2009). Solution Structure of a paradigm ArsR family zinc sensor in the DNA-bound state. PNAS 106:43 18177-18182.
- ↑ Chakravorty DK, Wang B, Lee CW, Giedroc DP, Merz KM Jr. Simulations of allosteric motions in the zinc sensor CzrA. J Am Chem Soc. 2012 Feb 22;134(7):3367-76. doi: 10.1021/ja208047b. Epub 2011 Nov , 14. PMID:22007899 doi:http://dx.doi.org/10.1021/ja208047b
- ↑ MacPherson S, Larochelle M, Turcotte B. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol Rev. 2006 Sep;70(3):583-604. PMID:16959962 doi:http://dx.doi.org/10.1128/MMBR.00015-06
- ↑ Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun 4;4(6):1609-1614.
- ↑ Grossoehme NE, Giedroc DP. Energetics of allosteric negative coupling in the zinc sensor S. aureus CzrA. J Am Chem Soc. 2009 Dec 16;131(49):17860-70. doi: 10.1021/ja906131b. PMID:19995076 doi:http://dx.doi.org/10.1021/ja906131b