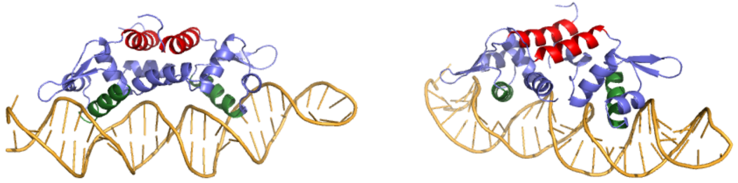Sandbox Reserved 1053
From Proteopedia
| Line 21: | Line 21: | ||
== DNA Binding == | == DNA Binding == | ||
| - | Ser 54, Ser 57, and His 58 are the primary sites of <scene name='69/694220/2kjb_colored/3'>DNA interaction</scene> in Czr A <ref name="critical"/>. These residues are likely to interact with the 5'-TGAA sequence found in the half-site of the DNA, where the alpha 4 helices (green) <scene name='69/694219/Czra_with_dna/2'>form an interaction with DNA</scene> (figure 3). | + | Ser 54, Ser 57, and His 58 are the primary sites of <scene name='69/694220/2kjb_colored/3'>DNA interaction</scene> in Czr A <ref name="critical"/>. These residues are likely to interact with the 5'-TGAA sequence found in the half-site of the DNA, where the alpha 4 helices (green) <scene name='69/694219/Czra_with_dna/2'>form an interaction with DNA</scene> (figure 3). Binding of two Zn <sup>+2</sup> ions <scene name='69/694220/Dna_residues_when_inhibited/2'>pushes these residues out of their DNA binding conformation</scene>. Additionally, Val 42 and Gln 53 (lime green) are involved in the <scene name='69/694220/Val_42_and_gln_53/1'>DNA binding pocket</scene>. This conclusion was experimentally determined by mutagenesis of the Gln and Val residues with an Ala and measuring the mutant DNA binding capacity. The DNA bound state of Czr A was tested by using the known critical residues for DNA interactions <ref name="critical"/>. <scene name='69/694220/Dna_binding_residues/2'>Critical DNA binding residues</scene> Gln 53, Val 42 (aqua), Ser 54, Ser 57, and His 58 (lime) were individually mutated to Ala, and kinetic experiments were performed. Compared to wild type Czr A, mutating Gln53 and V42 residues resulted in an 11-fold and 160-fold decrease in K<sub>a</sub>, respectively. Mutations to the main DNA interaction sites Ser 54, Ser 57, and His 58 result in binding similar to the inhibited non-DNA binding state, suggesting that these residues are essential to binding DNA. While the conformational change that occurs from the Zinc to DNA bound state of Czr A is small,the alpha 4 helices (shown in green in Figure 2) are slightly shifted. The loss of DNA binding in the mutagenesis experiements in combination with the lack of any other major physical changes between these two states further suggests that the alpha 4 helices are the location of DNA binding in Czr A. Experimental data can be found in table 1 from this same article. |
[[Image:800px-DNABound Final.fw.png CROPPED.fw.png|750px|thumb|center| Figure 3: Two views of Czr A bound to DNA. A segment of DNA is shown in orange with the alpha 5 helices displayed in red and the alpha 4 helices shown in green]] | [[Image:800px-DNABound Final.fw.png CROPPED.fw.png|750px|thumb|center| Figure 3: Two views of Czr A bound to DNA. A segment of DNA is shown in orange with the alpha 5 helices displayed in red and the alpha 4 helices shown in green]] | ||
Revision as of 15:46, 21 August 2017
Czr A: A Zinc Dependent Transcriptional Regulator
Background
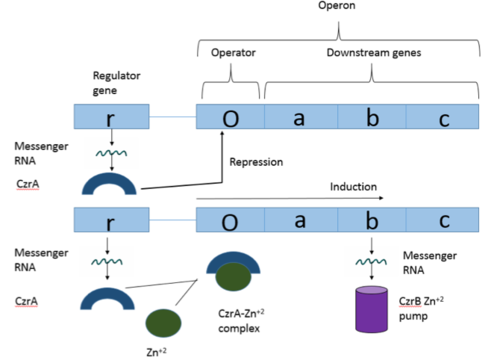
Operon Overview
Operons are a critical genetic component of most prokaryotic cells. There are many different operons, responsible for the production of proteins with a wide range of functions. The most well-known and studied operons are the Lac and Trp operons, responsible for producing enzymes which metabolize lactose and tryptophan respectively. Despite many differences in each operon and the proteins that they encode, operons all function in the same general manner (Figure 1). Structurally, each operon contains a regulator, an operator, and one or more structural genes. The regulator gene codes for a protein responsible for managing the expression level of the structural genes. The operator contains the binding sequence for RNA polymerase and is the site where transcription begins. Lastly, the structural genes code for proteins to be used elsewhere. The regulator protein (produced as a result of expression of the regulator gene) usually acts in a repressive manner. The regulator protein will bind to the operator gene, inhibiting the binding and/or progression of RNA polymerase to the structural genes, thus inhibiting transcription of the genes into mRNA. If the regulator protein were always active, the structural genes would never be expressed, so there must be a way to inactive the regulator protein, thus enabling expression of the structural genes. This is usually achieved through the binding of an inhibitor to the regulator protein. Since regulator proteins are DNA binding proteins, often this inhibition is allosteric rather than competitive. The inhibitor of the regulator protein binds to somewhere other than the active site of the protein, changing the regulator protein to decreases its affinity or ability to bind DNA and repress transcription.
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Arunkumar A., Campanello G., Giedroc D. (2009). Solution Structure of a paradigm ArsR family zinc sensor in the DNA-bound state. PNAS 106:43 18177-18182.
- ↑ MacPherson S, Larochelle M, Turcotte B. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol Rev. 2006 Sep;70(3):583-604. PMID:16959962 doi:http://dx.doi.org/10.1128/MMBR.00015-06
- ↑ Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun 4;4(6):1609-1614.
- ↑ Grossoehme NE, Giedroc DP. Energetics of allosteric negative coupling in the zinc sensor S. aureus CzrA. J Am Chem Soc. 2009 Dec 16;131(49):17860-70. doi: 10.1021/ja906131b. PMID:19995076 doi:http://dx.doi.org/10.1021/ja906131b