1wco
From Proteopedia
| Line 4: | Line 4: | ||
|PDB= 1wco |SIZE=350|CAPTION= <scene name='initialview01'>1wco</scene> | |PDB= 1wco |SIZE=350|CAPTION= <scene name='initialview01'>1wco</scene> | ||
|SITE= | |SITE= | ||
| - | |LIGAND= <scene name='pdbligand= | + | |LIGAND= <scene name='pdbligand=ABA:ALPHA-AMINOBUTYRIC+ACID'>ABA</scene>, <scene name='pdbligand=DAL:D-ALANINE'>DAL</scene>, <scene name='pdbligand=DBU:(2E)-2-AMINOBUT-2-ENOIC+ACID'>DBU</scene>, <scene name='pdbligand=DGL:D-GLUTAMIC+ACID'>DGL</scene>, <scene name='pdbligand=DHA:2-AMINO-ACRYLIC+ACID'>DHA</scene>, <scene name='pdbligand=FPP:FARNESYL+DIPHOSPHATE'>FPP</scene>, <scene name='pdbligand=MUB:N-ACETYLMURAMIC+ACID'>MUB</scene>, <scene name='pdbligand=NAG:N-ACETYL-D-GLUCOSAMINE'>NAG</scene> |
|ACTIVITY= | |ACTIVITY= | ||
|GENE= | |GENE= | ||
| + | |DOMAIN= | ||
| + | |RELATEDENTRY= | ||
| + | |RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1wco FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1wco OCA], [http://www.ebi.ac.uk/pdbsum/1wco PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=1wco RCSB]</span> | ||
}} | }} | ||
| Line 31: | Line 34: | ||
[[Category: Nuland, N A.J Van.]] | [[Category: Nuland, N A.J Van.]] | ||
[[Category: Tischenko, E.]] | [[Category: Tischenko, E.]] | ||
| - | [[Category: FPP]] | ||
| - | [[Category: MUB]] | ||
| - | [[Category: NAG]] | ||
[[Category: antibiotic]] | [[Category: antibiotic]] | ||
[[Category: peptidoglycan]] | [[Category: peptidoglycan]] | ||
| Line 39: | Line 39: | ||
[[Category: pyrophosphate cage]] | [[Category: pyrophosphate cage]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Mon Mar 31 00:34:25 2008'' |
Revision as of 21:34, 30 March 2008
| |||||||
| Ligands: | , , , , , , , | ||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
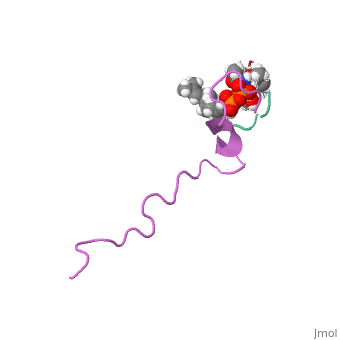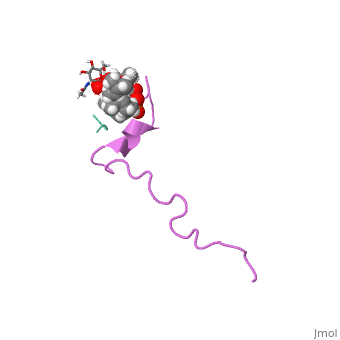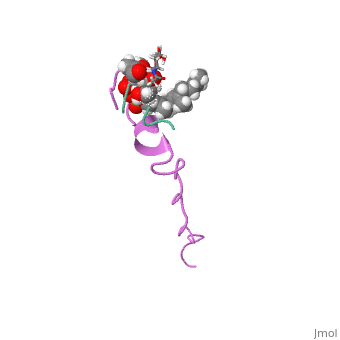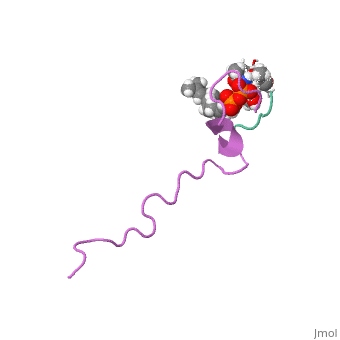
THE SOLUTION STRUCTURE OF THE NISIN-LIPID II COMPLEX
Overview
The emerging antibiotics-resistance problem has underlined the urgent need for novel antimicrobial agents. Lantibiotics (lanthionine-containing antibiotics) are promising candidates to alleviate this problem. Nisin, a member of this family, has a unique pore-forming activity against bacteria. It binds to lipid II, the essential precursor of cell wall synthesis. As a result, the membrane permeabilization activity of nisin is increased by three orders of magnitude. Here we report the solution structure of the complex of nisin and lipid II. The structure shows a novel lipid II-binding motif in which the pyrophosphate moiety of lipid II is primarily coordinated by the N-terminal backbone amides of nisin via intermolecular hydrogen bonds. This cage structure provides a rationale for the conservation of the lanthionine rings among several lipid II-binding lantibiotics. The structure of the pyrophosphate cage offers a template for structure-based design of novel antibiotics.
About this Structure
1WCO is a Single protein structure of sequence from Lactococcus lactis and Monarthropalpus flavus. This structure supersedes the now removed PDB entry 1UZT. Full crystallographic information is available from OCA.
Reference
The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics., Hsu ST, Breukink E, Tischenko E, Lutters MA, de Kruijff B, Kaptein R, Bonvin AM, van Nuland NA, Nat Struct Mol Biol. 2004 Oct;11(10):963-7. Epub 2004 Sep 12. PMID:15361862
Page seeded by OCA on Mon Mar 31 00:34:25 2008