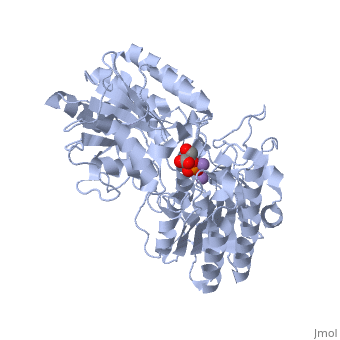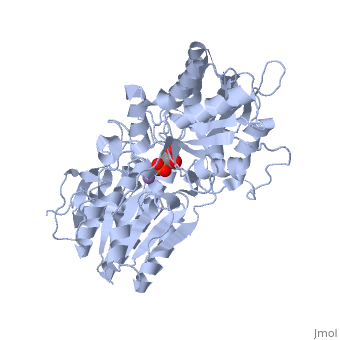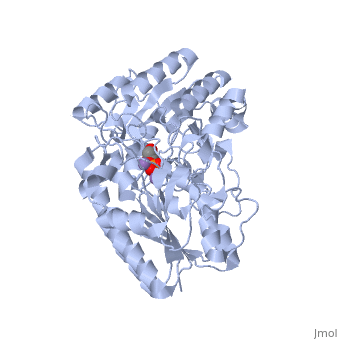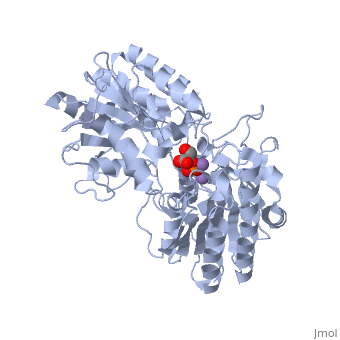
We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Phosphoglycerate Mutase
From Proteopedia
(Difference between revisions)
| Line 21: | Line 21: | ||
In order to understand how PGM catalyzes this reaction, an explanation of its active site is imperative. The most important residues in this enzyme include <scene name='40/401494/Cv/1'>His 8 and 181</scene> with imidazole groups which are in close proximity to carbons 2 and 3 in the substrate. His-8 is phosphorylated during during catalysis, and it is likely that His-179 acts as the proton donor/acceptor <ref>Rose, Z.B. (1980) Adv. Enzymol. Relat. Areas Mol. Biol. 51, 211-253</ref>. Based on crystallography experiments, the active site where these histidine residues reside lies at the bottom of a deep groove in each subunit. <ref name="winn" /> The sites in each subunit, whether the enzyme is a homodimer or homotetramer, are well separated. The active enzyme contains a phosphoryl group attached to His 8. This phosphoryl group is what is transferred to C2 of the substrate, resulting in an intermediate 2,3-bisphosphoglycerate-enzyme complex. Thus there is a <scene name='40/401494/Cv/3'>covalently attached phosphate</scene> in the active monomer. <ref name="voet" /> The phosphate group on C3 of the substrate is then transferred back onto His 8, thus regenerating the active form of the enzyme. | In order to understand how PGM catalyzes this reaction, an explanation of its active site is imperative. The most important residues in this enzyme include <scene name='40/401494/Cv/1'>His 8 and 181</scene> with imidazole groups which are in close proximity to carbons 2 and 3 in the substrate. His-8 is phosphorylated during during catalysis, and it is likely that His-179 acts as the proton donor/acceptor <ref>Rose, Z.B. (1980) Adv. Enzymol. Relat. Areas Mol. Biol. 51, 211-253</ref>. Based on crystallography experiments, the active site where these histidine residues reside lies at the bottom of a deep groove in each subunit. <ref name="winn" /> The sites in each subunit, whether the enzyme is a homodimer or homotetramer, are well separated. The active enzyme contains a phosphoryl group attached to His 8. This phosphoryl group is what is transferred to C2 of the substrate, resulting in an intermediate 2,3-bisphosphoglycerate-enzyme complex. Thus there is a <scene name='40/401494/Cv/3'>covalently attached phosphate</scene> in the active monomer. <ref name="voet" /> The phosphate group on C3 of the substrate is then transferred back onto His 8, thus regenerating the active form of the enzyme. | ||
| - | In addition to the importance of the two histidine residues in the active site, the amino acids that line the <scene name=' | + | In addition to the importance of the two histidine residues in the active site, the amino acids that line the <scene name='40/401494/Cv/4'>active site</scene> are also functionally important. These residues include H179, H8, E15, S11, T20, R59, and E86.<ref name="voet" /> Several positively charged residues line the active site pocket. These residues usually tend to be <scene name='40/401494/Cv/5'>arginine residues</scene>, which are important for the optimal activity of the enzyme. <ref name="winn" /> This structure is logical for its function because the enzyme binds a negatively charged substrate, thus a positively charged groove fosters tight binding with a negative substrate. The third and final important aspect of the active site is the presence of <scene name='40/401494/Cv/6'>glutamate residues 15 and 86</scene>.<ref name="winn" /> It is suggested that the carboxyl groups of these amino acid residues act as proton-withdrawing groups as they flank both sides of the substrate. |
== Kinetics == | == Kinetics == | ||
Revision as of 14:10, 31 August 2017
| |||||||||||
3D structures of phosphoglycerate mutase
Updated on 31-August-2017
Additional Resources
For additional information, please see: Carbohydrate Metabolism
References
- ↑ Crowhurst GS, Dalby AR, Isupov MN, Campbell JW, Littlechild JA. Structure of a phosphoglycerate mutase:3-phosphoglyceric acid complex at 1.7 A. Acta Crystallogr D Biol Crystallogr. 1999 Nov;55(Pt 11):1822-6. PMID:10531478
- ↑ http://disability.ucdavis.edu/disease_deatails.php?id=45
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 S., Winn I., Fothergill A. L., Harkins N. R., and Watson C. H. "Structure and Activity of Phosphoglycerate Mutase." Sciences 293.1063 (1981): 121-30. Print.
- ↑ "Phosphoglycerate mutase -." Wikipedia, the free encyclopedia. Web. 27 Feb. 2010. <http://en.wikipedia.org/wiki/Phosphoglycerate_mutase>.
- ↑ 5.0 5.1 5.2 Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry Life at the Molecular Level. New York: John Wiley & Sons, 2008. Print.
- ↑ Rose, Z.B. (1980) Adv. Enzymol. Relat. Areas Mol. Biol. 51, 211-253
- ↑ Rigden, D. J.; Walter, R. A.; Phillips, S. E. V.; Fothergill-Gilmore, L. A.Polyanionic inhibitors of phosphoglycerate mutase: combined structural and biochemical analysis J. Mol. Biol. 1999, 289, 691– 699
- ↑ McAleese, S.M., Fothergill-Gilmore, L.A.&Dixon, H.B.F. (1985) Biochem. J. 230, 535-542
- ↑ http://www.mda.org/disease/pgam.html
- ↑ http://disability.ucdavis.edu/disease_deatails.php?id=45
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Robert Trahin, Xuan Loi, David Canner, Christopher Vachon, Allie Paton