We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
SAM decarboxylase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
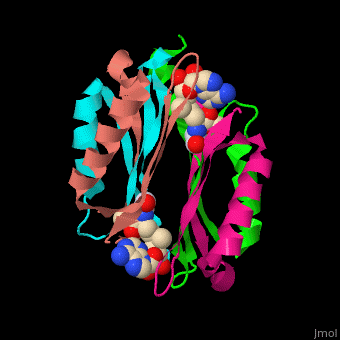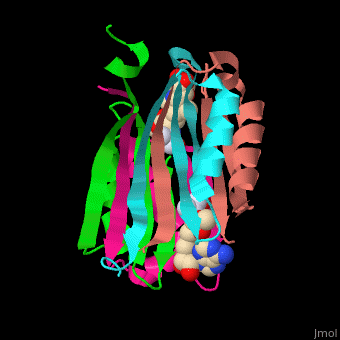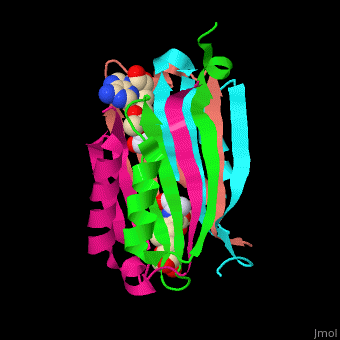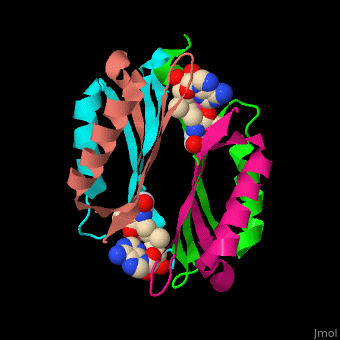
| - | <StructureSection load=' | + | <StructureSection load='' size='450' side='right' scene='49/493297/Cv/1' caption='S-adenosylmethionine decarboxylase with cofactor pyruvate complex with AdoMet [[3iwc]]'> |
== Function == | == Function == | ||
'''S-adenosylmethionine decarboxylase''' (AMD) catalyzes the conversion of S-adenosylmethionine (AdoMet) to S-adenosylmethioninamine . AMD is part of the polyamine biosynthesis, in particular in the biosynthesis of spermine and spermidine from putrescine. AMD uses a covalently bound pyruvate as a cofactor. The active AMD is generated by post-translational cleavage of a precursor molecule. The cleavage results in non-identical α and β subunits and the modification of a serine residue to pyruvate<ref>PMID:7948879</ref>. There are 2 classes of AMD. '''AMD I''' is found in bacteria and archae, '''AMD II''' is found in eukaryotes. | '''S-adenosylmethionine decarboxylase''' (AMD) catalyzes the conversion of S-adenosylmethionine (AdoMet) to S-adenosylmethioninamine . AMD is part of the polyamine biosynthesis, in particular in the biosynthesis of spermine and spermidine from putrescine. AMD uses a covalently bound pyruvate as a cofactor. The active AMD is generated by post-translational cleavage of a precursor molecule. The cleavage results in non-identical α and β subunits and the modification of a serine residue to pyruvate<ref>PMID:7948879</ref>. There are 2 classes of AMD. '''AMD I''' is found in bacteria and archae, '''AMD II''' is found in eukaryotes. | ||
Revision as of 05:41, 3 October 2017
| |||||||||||
3D structures of S-adenosylmethionine decarboxylase
Updated on 03-October-2017
References
- ↑ Mad Arif SA, Taylor MA, George LA, Butler AR, Burch LR, Davies HV, Stark MJ, Kumar A. Characterisation of the S-adenosylmethionine decarboxylase (SAMDC) gene of potato. Plant Mol Biol. 1994 Oct;26(1):327-38. PMID:7948879
- ↑ Weisel FC, Kloepping C, Pichl A, Sydykov A, Kojonazarov B, Wilhelm J, Roth M, Ridge KM, Igarashi K, Nishimura K, Maison W, Wackendorff C, Klepetko W, Jaksch P, Ghofrani HA, Grimminger F, Seeger W, Schermuly RT, Weissmann N, Kwapiszewska G. Impact of S-adenosylmethionine decarboxylase 1 on pulmonary vascular remodeling. Circulation. 2014 Apr 8;129(14):1510-23. doi: 10.1161/CIRCULATIONAHA.113.006402. , Epub 2014 Jan 27. PMID:24470481 doi:http://dx.doi.org/10.1161/CIRCULATIONAHA.113.006402
- ↑ Bale S, Baba K, McCloskey DE, Pegg AE, Ealick SE. Complexes of Thermotoga maritimaS-adenosylmethionine decarboxylase provide insights into substrate specificity. Acta Crystallogr D Biol Crystallogr. 2010 Feb;66(Pt 2):181-9. Epub 2010, Jan 22. PMID:20124698 doi:10.1107/S090744490904877X
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Jaime Prilusky