We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Alec Bertsch/sandbox 1
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
==L5/5S rRNA Complex== | ==L5/5S rRNA Complex== | ||
<StructureSection load='1mji' size='340' side='right' caption='Caption for this structure' scene=''> | <StructureSection load='1mji' size='340' side='right' caption='Caption for this structure' scene=''> | ||
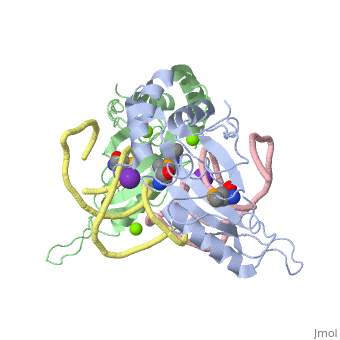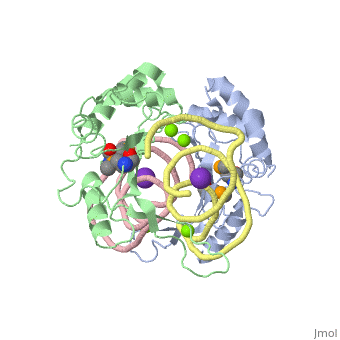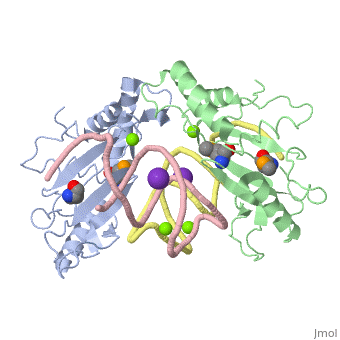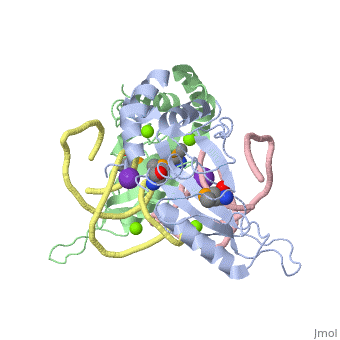
| - | The ribosome is an essential component of all living cells. It is responsible for translation of proteins using messenger RNA (mRNA) and transfer RNA (tRNA). Ribosomes are composed of both proteins and RNA known as ribosomal RNA (rRNA). There are three functional sites found within the ribosome that are essential for translation: the A, P, and E-sites. The A-site is responsible for recruiting the tRNA and the E-site is where the tRNA leaves from once the protein is removed in the P-site. The site of interest for this sandbox is the P-site, where the protein is removed from the tRNA and added to the growing chain of amino acids. Its primary component is the L5/5S rRNA complex. | + | The ribosome is an essential component of all living cells. It is responsible for translation of proteins using messenger RNA (mRNA) and transfer RNA (tRNA). Ribosomes are composed of both proteins and RNA known as ribosomal RNA (rRNA). There are three functional sites found within the ribosome that are essential for translation: the A, P, and E-sites. The A-site is responsible for recruiting the tRNA and the E-site is where the tRNA leaves from once the protein is removed in the P-site. The site of interest for this sandbox is the P-site, where the protein is removed from the tRNA and added to the growing chain of amino acids. Its primary component is the L5/5S rRNA complex. This complex has been proven to be an essential component to both the structure and the function of the ribosome. |
This is a default text for your page '''Alec Bertsch/sandbox 1'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | This is a default text for your page '''Alec Bertsch/sandbox 1'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
Revision as of 01:08, 10 October 2017
L5/5S rRNA Complex
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644