User:Alec Bertsch/sandbox 1
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
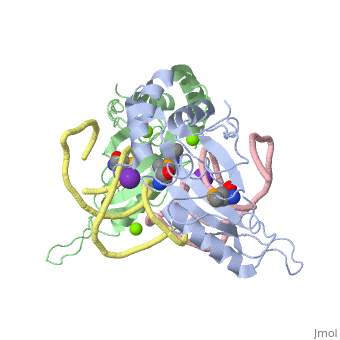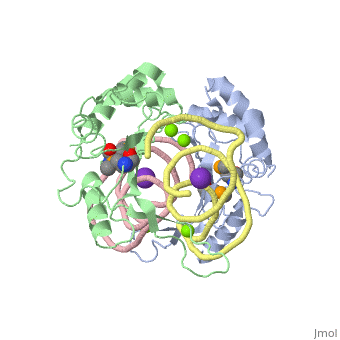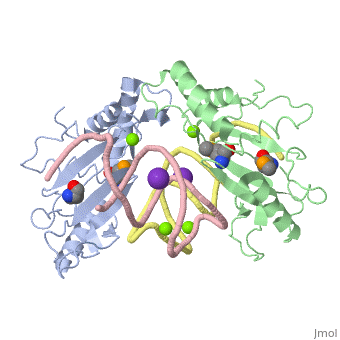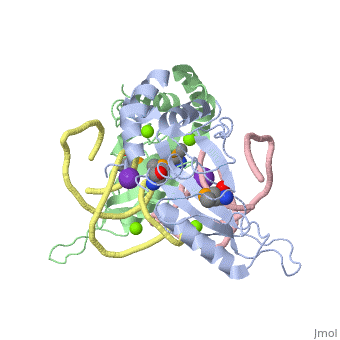
The complex is responsible for the stabilization of the ribosomal proteins, mRNA, and tRNA, and is crucial for protein synthesis to occur. The primary function of the L5 protein is to anchor the peptide-linked tRNA to the P-site for the transfer of the peptide to the growing chain of amino acids.<ref>PMID:11497428</ref> | The complex is responsible for the stabilization of the ribosomal proteins, mRNA, and tRNA, and is crucial for protein synthesis to occur. The primary function of the L5 protein is to anchor the peptide-linked tRNA to the P-site for the transfer of the peptide to the growing chain of amino acids.<ref>PMID:11497428</ref> | ||
The L5 protein is also responsible for the assembly of the ribosome. In ribosomes that fail to synthesize the L5 protein, the ribosome is unable to connect the large and small sub-units together. Without this protein, the cell would only be able to divide a select few times.<ref>doi:10.1093/nar/gks676</ref> | The L5 protein is also responsible for the assembly of the ribosome. In ribosomes that fail to synthesize the L5 protein, the ribosome is unable to connect the large and small sub-units together. Without this protein, the cell would only be able to divide a select few times.<ref>doi:10.1093/nar/gks676</ref> | ||
| - | The 5S rRNA is responsible for enhancing protein synthesis. This is done by stabilizing the structure of the ribosome through its tight binding to the L5 protein.<ref>PMID:11440169</ref> 5S rRNA has also been noted to be a major component in tumor suppression. This occurs when this rRNA joins with other molecules to form the 5S ribonucleoprotein particle (RNP) which accumlates when ribosome biogenesis is suppressed or blocked.<ref>PMID:27528756</ref> | + | The 5S rRNA is responsible for enhancing protein synthesis. This is done by stabilizing the structure of the ribosome through its tight binding to the L5 protein.<ref>PMID:11440169</ref> 5S rRNA has also been noted to be a major component in tumor suppression. This occurs when this rRNA joins with other molecules to form the 5S ribonucleoprotein particle (RNP) which accumlates when ribosome biogenesis is suppressed or blocked.<ref>PMID:27528756</ref> The extent of its role in ribosomal function is not well known, but there have been studies to suggest that the 5S rRNA helps with the binding of the tRNA within the P-site.<ref>PMID:8722002</ref> |
== Structural highlights == | == Structural highlights == | ||
Revision as of 12:22, 10 October 2017
L5/5S rRNA Complex
| |||||||||||
References
- ↑ Wise JP, Leonard JC, Patierno SR. Clastogenicity of lead chromate particles in hamster and human cells. Mutat Res. 1992 Jan;278(1):69-79. PMID:1370121
- ↑ Meskauskas A, Dinman JD. Ribosomal protein L5 helps anchor peptidyl-tRNA to the P-site in Saccharomyces cerevisiae. RNA. 2001 Aug;7(8):1084-96. PMID:11497428
- ↑ Korepanov AP, Korobeinikova AV, Shestakov SA, Garber MB, Gongadze GM. Protein L5 is crucial for in vivo assembly of the bacterial 50S ribosomal subunit central protuberance. Nucleic Acids Res. 2012 Oct;40(18):9153-9. doi: 10.1093/nar/gks676. Epub 2012 Jul, 20. PMID:22821559 doi:http://dx.doi.org/10.1093/nar/gks676
- ↑ Barciszewska MZ, Szymanski M, Erdmann VA, Barciszewski J. Structure and functions of 5S rRNA. Acta Biochim Pol. 2001;48(1):191-8. PMID:11440169
- ↑ Pelava A, Schneider C, Watkins NJ. The importance of ribosome production, and the 5S RNP-MDM2 pathway, in health and disease. Biochem Soc Trans. 2016 Aug 15;44(4):1086-90. doi: 10.1042/BST20160106. PMID:27528756 doi:http://dx.doi.org/10.1042/BST20160106
- ↑ Bogdanov AA, Dontsova OA, Dokudovskaya SS, Lavrik IN. Structure and function of 5S rRNA in the ribosome. Biochem Cell Biol. 1995 Nov-Dec;73(11-12):869-76. PMID:8722002
- ↑ Wise JP, Leonard JC, Patierno SR. Clastogenicity of lead chromate particles in hamster and human cells. Mutat Res. 1992 Jan;278(1):69-79. PMID:1370121
- ↑ Wise JP, Leonard JC, Patierno SR. Clastogenicity of lead chromate particles in hamster and human cells. Mutat Res. 1992 Jan;278(1):69-79. PMID:1370121
- ↑ doi: https://dx.doi.org/10.1017/S1355838202029953