1ygp
From Proteopedia
| Line 4: | Line 4: | ||
|PDB= 1ygp |SIZE=350|CAPTION= <scene name='initialview01'>1ygp</scene>, resolution 2.8Å | |PDB= 1ygp |SIZE=350|CAPTION= <scene name='initialview01'>1ygp</scene>, resolution 2.8Å | ||
|SITE= | |SITE= | ||
| - | |LIGAND= <scene name='pdbligand= | + | |LIGAND= <scene name='pdbligand=PLP:PYRIDOXAL-5'-PHOSPHATE'>PLP</scene>, <scene name='pdbligand=PO4:PHOSPHATE+ION'>PO4</scene> |
|ACTIVITY= | |ACTIVITY= | ||
|GENE= YEAST GLYCOGEN PHOSPHORYLASE ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=4932 Saccharomyces cerevisiae]) | |GENE= YEAST GLYCOGEN PHOSPHORYLASE ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=4932 Saccharomyces cerevisiae]) | ||
| + | |DOMAIN= | ||
| + | |RELATEDENTRY= | ||
| + | |RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1ygp FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1ygp OCA], [http://www.ebi.ac.uk/pdbsum/1ygp PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=1ygp RCSB]</span> | ||
}} | }} | ||
| Line 28: | Line 31: | ||
[[Category: Lin, K.]] | [[Category: Lin, K.]] | ||
[[Category: Rath, V L.]] | [[Category: Rath, V L.]] | ||
| - | [[Category: PLP]] | ||
| - | [[Category: PO4]] | ||
[[Category: glycosyltransferase]] | [[Category: glycosyltransferase]] | ||
[[Category: phosphorylated form]] | [[Category: phosphorylated form]] | ||
[[Category: yeast]] | [[Category: yeast]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Mon Mar 31 01:06:46 2008'' |
Revision as of 22:06, 30 March 2008
| |||||||
| , resolution 2.8Å | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||
| Gene: | YEAST GLYCOGEN PHOSPHORYLASE (Saccharomyces cerevisiae) | ||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
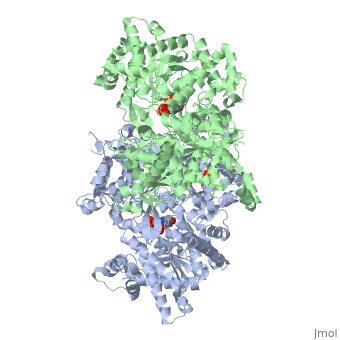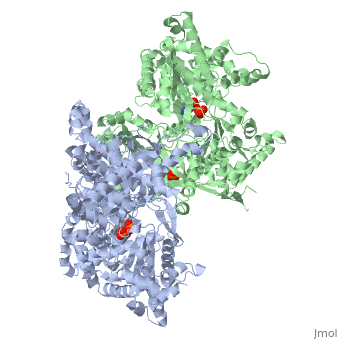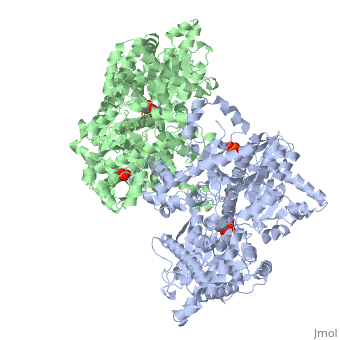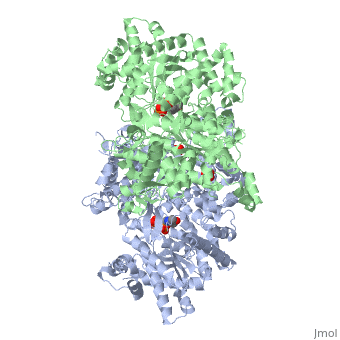
PHOSPHORYLATED FORM OF YEAST GLYCOGEN PHOSPHORYLASE WITH PHOSPHATE BOUND IN THE ACTIVE SITE.
Overview
A phosphorylation-initiated mechanism of local protein refolding activates yeast glycogen phosphorylase (GP). Refolding of the phosphorylated amino-terminus was shown to create a hydrophobic cluster that wedges into the subunit interface of the enzyme to trigger activation. The phosphorylated threonine is buried in the allosteric site. The mechanism implicates glucose 6-phosphate, the allosteric inhibitor, in facilitating dephosphorylation by dislodging the buried covalent phosphate through binding competition. Thus, protein phosphorylation-dephosphorylation may also be controlled through regulation of the accessibility of the phosphorylation site to kinases and phosphatases. In mammalian glycogen phosphorylase, phosphorylation occurs at a distinct locus. The corresponding allosteric site binds a ligand activator, adenosine monophosphate, which triggers activation by a mechanism analogous to that of phosphorylation in the yeast enzyme.
About this Structure
1YGP is a Single protein structure of sequence from Saccharomyces cerevisiae. The following page contains interesting information on the relation of 1YGP with [Glycogen Phosphorylase]. Full crystallographic information is available from OCA.
Reference
A protein phosphorylation switch at the conserved allosteric site in GP., Lin K, Rath VL, Dai SC, Fletterick RJ, Hwang PK, Science. 1996 Sep 13;273(5281):1539-42. PMID:8703213
Page seeded by OCA on Mon Mar 31 01:06:46 2008