We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Glycogen Phosphorylase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
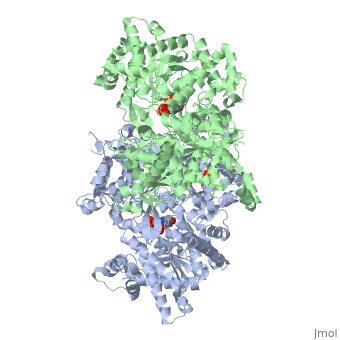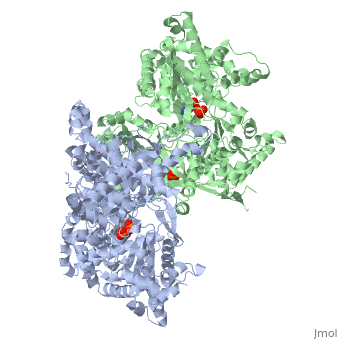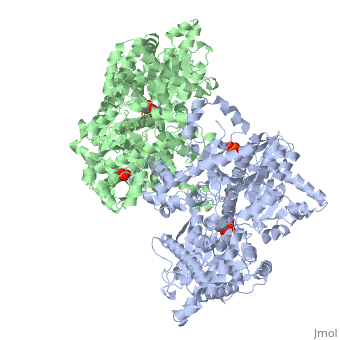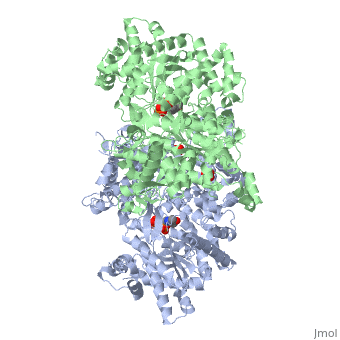
| + | <StructureSection load='1ygp' size='350' side='right' caption='Yeast glycogen phosphorylase dimer with pyridoxal-5-phosphate and phosphate (PDB entry [[1ygp]])' scene=''> | ||
=Introduction= | =Introduction= | ||
'''Glycogen phosphorylase''' catalyzes the hydrolysis of glycogen to generate glucose-1-phosphate and shortened glycogen molecule and is considered the rate limiting step in the degradation of glycogen<ref name="gp">PMID: 15214781 </ref>. It is a part of the glucosyltransferase family and acts on the α-1,4-glycosidic linkage; the phosphorylase comes to a standstill 4 residues from an α-1,6-branchpoint, where debranching enzyme takes over <ref name =“gp3”> PMID: 11949930</ref>. The glucose-1-phophate is then further degraded via the pathway of glycolysis. Studies have found that mammals have liver, muscle and brain isoforms of phosphorylase but it is found among all species; muscle glycogen phosphorylase is present to degrade glycogen to forms of energy by means of glycolysis during muscle contractions and liver glycogen is present to regulate the blood glucose levels within the blood <ref name =“gp3”/><ref name="PLP">Palm D, Klein HW, Schinzel R, Buehner M, Helmreich EJM. The role of pyridoxal 5’-phosphate in glycogen phosphorylase catalysis. Biochemistry. 1990 Feb 6; 29(5):1099-1107.</ref>. See also [[Glycogen Metabolism & Gluconeogenesis]]. | '''Glycogen phosphorylase''' catalyzes the hydrolysis of glycogen to generate glucose-1-phosphate and shortened glycogen molecule and is considered the rate limiting step in the degradation of glycogen<ref name="gp">PMID: 15214781 </ref>. It is a part of the glucosyltransferase family and acts on the α-1,4-glycosidic linkage; the phosphorylase comes to a standstill 4 residues from an α-1,6-branchpoint, where debranching enzyme takes over <ref name =“gp3”> PMID: 11949930</ref>. The glucose-1-phophate is then further degraded via the pathway of glycolysis. Studies have found that mammals have liver, muscle and brain isoforms of phosphorylase but it is found among all species; muscle glycogen phosphorylase is present to degrade glycogen to forms of energy by means of glycolysis during muscle contractions and liver glycogen is present to regulate the blood glucose levels within the blood <ref name =“gp3”/><ref name="PLP">Palm D, Klein HW, Schinzel R, Buehner M, Helmreich EJM. The role of pyridoxal 5’-phosphate in glycogen phosphorylase catalysis. Biochemistry. 1990 Feb 6; 29(5):1099-1107.</ref>. See also [[Glycogen Metabolism & Gluconeogenesis]]. | ||
| - | |||
| - | <StructureSection load='1ygp' size='350' side='right' caption='Yeast glycogen phosphorylase dimer with pyridoxal-5-phosphate and phosphate (PDB entry [[1ygp]])' scene=''> | ||
| - | |||
| - | |||
=Structure and Function= | =Structure and Function= | ||
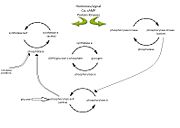
Glycogen phosphorylase is a dimer consisting of two identical subunits and has an essential cofactor, <scene name='Sandbox_153/Plp/1'>pryridoxal phosphate (PLP)</scene><ref name="PLP"/>. Glycogen phosphorylase can be found in two different states, glycogen phosphorylase a (GP''a'') and glycogen phosphorylase b (GP''b'')<ref name="gp"/>The difference in the structures is due to phosphorylation of the <scene name='Sandbox_153/Ser14/1'>Ser-14</scene> residue which results in the active form (GP''a''). Protein phosphatases dephosphorylate the GP''a'' to the inactive form, also known as GP''b''. Both forms of glycogen phosphorylase can also be found in T and R states where T is the inactive state because it appears to have a low affinity for substrate and R is the active state where it appears to have a greater affinity for substrate<ref name="gp2">PMID: 1900534</ref>. | Glycogen phosphorylase is a dimer consisting of two identical subunits and has an essential cofactor, <scene name='Sandbox_153/Plp/1'>pryridoxal phosphate (PLP)</scene><ref name="PLP"/>. Glycogen phosphorylase can be found in two different states, glycogen phosphorylase a (GP''a'') and glycogen phosphorylase b (GP''b'')<ref name="gp"/>The difference in the structures is due to phosphorylation of the <scene name='Sandbox_153/Ser14/1'>Ser-14</scene> residue which results in the active form (GP''a''). Protein phosphatases dephosphorylate the GP''a'' to the inactive form, also known as GP''b''. Both forms of glycogen phosphorylase can also be found in T and R states where T is the inactive state because it appears to have a low affinity for substrate and R is the active state where it appears to have a greater affinity for substrate<ref name="gp2">PMID: 1900534</ref>. | ||
Revision as of 08:59, 20 October 2017
| |||||||||||
3D structures of glycogen phosphorylase
Updated on 20-October-2017
Additional Resources
For additional information, see: Carbohydrate Metabolism
References
- ↑ 1.0 1.1 1.2 Kristiansen M, Andersen B, Iversen LF, Westergaard N. Identification, synthesis, and characterization of new glycogen phosphorylase inhibitors binding to the allosteric AMP site. J Med Chem. 2004 Jul 1;47(14):3537-45. PMID:15214781 doi:10.1021/jm031121n
- ↑ 2.0 2.1 Roach PJ. Glycogen and its metabolism. Curr Mol Med. 2002 Mar;2(2):101-20. PMID:11949930
- ↑ 3.0 3.1 3.2 3.3 Palm D, Klein HW, Schinzel R, Buehner M, Helmreich EJM. The role of pyridoxal 5’-phosphate in glycogen phosphorylase catalysis. Biochemistry. 1990 Feb 6; 29(5):1099-1107.
- ↑ 4.0 4.1 4.2 4.3 4.4 Barford D, Hu SH, Johnson LN. Structural mechanism for glycogen phosphorylase control by phosphorylation and AMP. J Mol Biol. 1991 Mar 5;218(1):233-60. PMID:1900534
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 5.7 Fletterick RJ, Sprang SR. Glycogen phosphorylase Structures and function. Accounts of Chemical Research. 1982 Nov; 15(11):361-369.
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 6.6 6.7 Johnson LH. Glycogen Phosphorylase: Control by phosphorylation and allosteric effectors. The FASEB Journal. 1992 March;6:2274-2282.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Amy Chahal, Ann Taylor, Alexander Berchansky, Joel L. Sussman, Riley Hicks, Andrea Gorrell, David Canner