We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Journal:Structure:1
From Proteopedia
(Difference between revisions)

| Line 6: | Line 6: | ||
Protein-protein interactions (PPI) mediate most major processes in the cell. Despite the crowded cellular environment, proteins maintain a high degree of specificity in their interactions. For this, proteins evolved to balance between the ability to bind the desired partners while rejecting all other proteins. | Protein-protein interactions (PPI) mediate most major processes in the cell. Despite the crowded cellular environment, proteins maintain a high degree of specificity in their interactions. For this, proteins evolved to balance between the ability to bind the desired partners while rejecting all other proteins. | ||
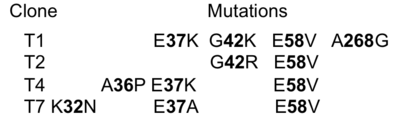
| - | Here, we address the question of the sequence distance to generate new binding. In other words, how many mutations have to be inserted into a protein so that it will bind a given partner. For this we generated a random library of TEM1- | + | Here, we address the question of the sequence distance to generate new binding. In other words, how many mutations have to be inserted into a protein so that it will bind a given partner. For this we generated a random library of TEM1-β-lactamase mutant proteins displayed on yeast and selected the library to bind TEM1 wild-type. We found that three mutations were sufficient to develop a <scene name='76/763767/Cv/2'>de-novo protein interaction</scene> (see Table below). The position of each of the selected mutations is represented as orange sticks on the TEM1-WT scaffold (PDB [[1jtg]] chain A). |
{{Clear}} | {{Clear}} | ||
[[Image:Gid1.png|thumb|The identities of the mutations present in the 4 selected clones are provided in the table|400px|left]] | [[Image:Gid1.png|thumb|The identities of the mutations present in the 4 selected clones are provided in the table|400px|left]] | ||
Revision as of 07:11, 24 October 2017
| |||||||||||
- ↑ REF
- ↑ Reichmann D, Cohen M, Abramovich R, Dym O, Lim D, Strynadka NC, Schreiber G. Binding hot spots in the TEM1-BLIP interface in light of its modular architecture. J Mol Biol. 2007 Jan 19;365(3):663-79. Epub 2006 Oct 3. PMID:17070843 doi:10.1016/j.jmb.2006.09.076
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.