User:Brittany Allen/Sandbox 1
From Proteopedia
| Line 6: | Line 6: | ||
== Structure == | == Structure == | ||
TIP30 is an evolutionary conserved gene located on human chromosome 11<ref>1</ref> . TIP30 has a molecular mass of 30 kd<ref>6</ref> and is composed of 242 amino acids<ref> 1 </ref>. TIP30 is composed of alpha helices, beta sheets, and loops (Movie). Through sequence analysis studies, it is thought that TIP30 may be a member of the SDR (substrate determining residue) family which contain a characteristic motif at their catalytic start sites<ref> 1 </ref>. The carboxyl terminus of TIP30 binds to the SDR substrate, while the amino terminus of TIP30 is the nucleotide cofactor-binding domain which has a characteristic Gly-X-X-Gly- X-X-Gly motif (where X can be any amino acid)<ref> 1 </ref>. Since SDR families have binding specificity for NADPH<ref> 8 </ref> and TIP30 contains a dehydrogenase reductase fold that contains binding specificity for NADPH<ref> 1 </ref>., the binding of NADPH may be important for the biological activity of TIP30 including interactions with importins as well as the c-myc system<ref> 8 </ref>. | TIP30 is an evolutionary conserved gene located on human chromosome 11<ref>1</ref> . TIP30 has a molecular mass of 30 kd<ref>6</ref> and is composed of 242 amino acids<ref> 1 </ref>. TIP30 is composed of alpha helices, beta sheets, and loops (Movie). Through sequence analysis studies, it is thought that TIP30 may be a member of the SDR (substrate determining residue) family which contain a characteristic motif at their catalytic start sites<ref> 1 </ref>. The carboxyl terminus of TIP30 binds to the SDR substrate, while the amino terminus of TIP30 is the nucleotide cofactor-binding domain which has a characteristic Gly-X-X-Gly- X-X-Gly motif (where X can be any amino acid)<ref> 1 </ref>. Since SDR families have binding specificity for NADPH<ref> 8 </ref> and TIP30 contains a dehydrogenase reductase fold that contains binding specificity for NADPH<ref> 1 </ref>., the binding of NADPH may be important for the biological activity of TIP30 including interactions with importins as well as the c-myc system<ref> 8 </ref>. | ||
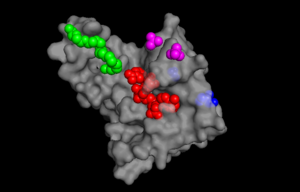
| - | [[Image: pymol ligand pic.png |300px|left|thumb| Figure 1. The binding sites of TIP30 as identified using PYMOL.]] | ||
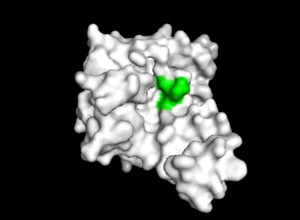
| - | TIP30 is known to bind 4 ligands: NDP (NADPH Dihydro-nicotinamide-adenine-dinucleotide-phosphate), PE8 (3,6,9,12,15,18,21 heptatricosane-1,2,3-diol), GOL (glycerol), and SO4 (sulfate ion)<ref> 9 </ref>. The location these residues bind can be seen in ________. According to uniprot, TIP30 contains a nucleotide binding region between residues 19-52, as shown in ________, and a binding site at residue 131, as shown in______. When comparing _____ and ________, it can be observed that the ligands bind in the active regions and interact with the residues. Uniprot also noted that TIP30 can contain a mutagenic site at positions 28-31 (______), if this site is present there is a loss of proapoptotic and metastasis-inhibiting effects. | ||
| - | [[Image: resi19-52 site.png |300px|right|thumb| | + | TIP30 is known to bind 4 ligands: NDP (NADPH Dihydro-nicotinamide-adenine-dinucleotide-phosphate), PE8 (3,6,9,12,15,18,21 heptatricosane-1,2,3-diol), GOL (glycerol), and SO4 (sulfate ion)<ref> 9 </ref>. The location these residues bind can be seen in image 1. [[Image: pymol ligand pic.png |300px|left|thumb| Image 1. The binding sites of TIP30 as identified using PYMOL.]] According to Uniprot, TIP30 contains a nucleotide binding region between residues 19-52, as shown in image 2. [[Image: resi19-52 site.png |300px|right|thumb| Image 2. TIP30 displayed showing residues 19-52.]] and a binding site at residue 131. When comparing images 1 and 2, it can be observed that the ligands bind in the active regions and interact with the residues. Uniprot also noted that TIP30 can contain a mutagenic site at positions 28-31 (image 3), [[Image: resi 28-31 mutagenesis.png |300px|left|thumb| Image 3. Mutagenesis]]if this site is present there is a loss of proapoptotic and metastasis-inhibiting effects. |
| + | |||
| + | |||
| + | |||
| - | [[Image: resi 28-31 mutagenesis.png |300px|left|thumb| Figure 3. Mutagenesis]] | ||
== Function == | == Function == | ||
Revision as of 02:47, 4 December 2017
|
Contents |
General Background Information
TIP30, also known as both CC3[1]. TIP30 acts as a transcription cofactor that can regulate gene expression[2] and has both pro-apoptotic and anti-metastatic properties[3]. When the promoter of TIP30 is methylated, TIP30 becomes downregulated and associated with tumor prognosis[4]. It is thought that the tumor suppressor effect is the result of the inhibition of nuclear transport through binding with importin βs or by regulating transcription through interaction as a complex with a co-activator independent of AF-2 function and the c-myc gene[5]. Several studies have found that TIP30 may be linked to esophageal carcinoma, laryngeal carcinoma, glioma, pancreatic ductal adenocarcinoma, breast cancer, gastric cancer, gallbladder adenocarcinoma, lung cancer, and hepatocellular carcinoma[6].
Structure
TIP30 is an evolutionary conserved gene located on human chromosome 11[7] . TIP30 has a molecular mass of 30 kd[8] and is composed of 242 amino acids[9]. TIP30 is composed of alpha helices, beta sheets, and loops (Movie). Through sequence analysis studies, it is thought that TIP30 may be a member of the SDR (substrate determining residue) family which contain a characteristic motif at their catalytic start sites[10]. The carboxyl terminus of TIP30 binds to the SDR substrate, while the amino terminus of TIP30 is the nucleotide cofactor-binding domain which has a characteristic Gly-X-X-Gly- X-X-Gly motif (where X can be any amino acid)[11]. Since SDR families have binding specificity for NADPH[12] and TIP30 contains a dehydrogenase reductase fold that contains binding specificity for NADPH[13]., the binding of NADPH may be important for the biological activity of TIP30 including interactions with importins as well as the c-myc system[14].
TIP30 is known to bind 4 ligands: NDP (NADPH Dihydro-nicotinamide-adenine-dinucleotide-phosphate), PE8 (3,6,9,12,15,18,21 heptatricosane-1,2,3-diol), GOL (glycerol), and SO4 (sulfate ion)[15]. The location these residues bind can be seen in image 1. According to Uniprot, TIP30 contains a nucleotide binding region between residues 19-52, as shown in image 2. and a binding site at residue 131. When comparing images 1 and 2, it can be observed that the ligands bind in the active regions and interact with the residues. Uniprot also noted that TIP30 can contain a mutagenic site at positions 28-31 (image 3), if this site is present there is a loss of proapoptotic and metastasis-inhibiting effects.