Pro-protein convertase subtilisin/kexin type 9 (PCSK9) the ninth known member of the mammalian serine proprotein convertase (PC) family, and plays an important role in low density lipoproteins (LDL) metabolism. Once secreted, PCSK9 binds LDL receptors (LDLRs), targeting them toward intracellular degradation through an endosomal/lysosomal route. Inhibition of PCSK9 can reduce LDLRs degradation and increase the expression of LDLRs in the cell surface, resulting in an enhanced recycling of LDLRs and a reduction in the levels of LDL cholesterol. Hence, inhibitors of PCSK9 suppose a promising therapeutic strategy for the treatment of hypercholesterolemia.
Discovery of PCSK9
PCSK9 was first described as neural apoptosis-regulated convertase 1 (NARC-1) in studies of cerebral neuron apoptosis, suggesting that it could be implicated in the differentiation of cortical neurons [1]. Concomitant and following studies in patients with familiar hypercholesterolemia revealed the clinical importance of PCSK9, showing that patients with gain-of-function mutations presented increased levels of cholesterol in plasma (i.e. hypercholesterolemia) due to reduced expression of LDLRs. In contrast, loss-of-function variants of PSCK9 are associated with a reduction of LDL cholesterol levels and a lower risk of cardiovascular disease. The role of PSCK9 in LDLRs and cholesterol metabolism has been confirmed in animal models. Thus, mice overexpressing PCSK9 show a reduction in the expression of hepatic LDLRs and hypercholesterolemia, whereas knockout mice for PCSK9 present decreased levels of plasmatic LDL cholesterol because of increased expression of LDLRs [2][3].
Gene expression and synthesis of PCSK9
PCSK9 is the ninth known member of the mammalian subtilisin (S8) serine proprotein convertase (PC) family that carries out the proteolytic maturation of secretory proteins such as neuropeptides, prohormones and cytokines. Humans have nine different PCs that can be divided between S8A and S8B subfamilies. PCSK9 is classified in subfamily S8A [4].
The human gene for PCSK9 is 22 kb length and it is located in chromosome 1p32.3. It contains 11 introns and 12 exons that encode the 692 amino acids of the enzyme. The sequence of the protein is characterized by a signal sequence (amino acids 1-30), a prodomain (amino acids 31-152), and a catalytic domain, followed by a C-terminal region of 243 amino acids which is rich in cysteine and histidine residues. PCSK9 is mainly expressed in the liver, intestine and kidney, and it can also be in the nervous system. It is synthesized as a precursor of ~74 kDa that is processed in the endoplasmic reticulum (ER) where it undergoes cleavage of its signal peptide and intramolecular autocatalytic cleavage producing a ~60-kDa catalytic fragment. The autocatalysis of the zymogen takes place between Gln152 and Ser153
[5]. This cleavage is necessary for transport from ER to the Golgi body and for secretion. The cleaved prodomain of ~14 kDa remains associated with the catalytic domain, which is unique to PCSK9. This facilitates protein folding, permits the mature protein to move from ER into the secretory pathway and regulates the catalytic activity of the enzyme by blocking the access to the catalytic site
[2][3].
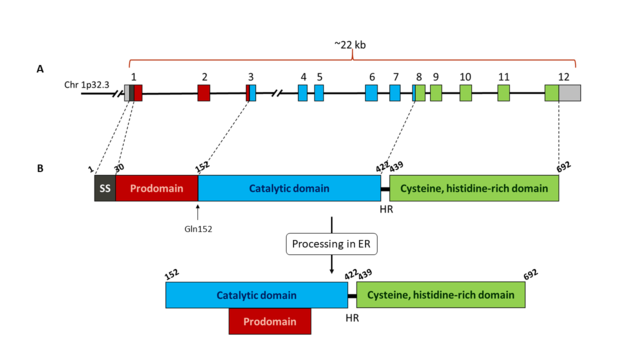
Schematic representation of PCSK9 gene (A) and protein (B). A: exons of PCSK9 gene are shown as coloured boxes. Each colour corresponds to a different domain of the protein. B: Representation of PCSK9 protein domains: signal sequence (SS, black), prodomain (red), catalytic domain (blue) and Cys, His rich C-terminal domain (green). The autocatalytic cleavage site at Gln152 is indicated with an arrow. Once the protein has been processed in the endoplasmic reticulum (ER), the prodomain remains bound to the catalytic domain. The catalytic domain is linked to the C-terminal domain through a 18 amino acids hinge region (HR). Numbers above each domain indicate the amino acid number of the protein sequence.
PCSK9 can be found in plasma in two forms: the mature and secreted form of ~60 kDa, and as an inactivated fragment of ~53 kDa produced by the cleavage of the mature form at the motive RFHR218↓ by other proprotein convertases, mainly furin and/or PC5/6A [6].
In humans, PCSK9 circulates in plasma in a phosphorylated state and it has been shown that it is phosphorylated at the Ser47 and Ser688 by a Golgi casein kinase-like kinase ex vivo. This phosphorylation might be important to protect the propeptide against proteolysis [7].
Regulation of PCSK9 gene expression
The expression of PCSK9 and LDLR genes are regulated by a common pathway. Thus, when there is reduction in the cholesterol cell content, the transcription of both genes is induced through the sterol regulatory element-binding protein (SREBP) [8]. Additionally, there are evidences that statins also upregulate the expression of PCSK9 gene [9].
Another factor that affects the expression of PCSK9 is the nutritional status. It has been shown that fasting decreases the expression of hepatic PCSK9 in mice, and that the level of PCSK9 gene expression is recovered upon refeeding. Furthermore, insulin also upregulates the hepatic PCSK9 gene expression, suggesting a possible role of PCSK9 in insulin-related diseases like type 2 diabetes. The liver X receptor (LXR) and sterol regulatory element-binding protein 1c (SREBP-1c) would be involved in the pathway that regulates the expression of PCSK9 according to the nutritional status [8].
Function
Although the first role suggested for PCSK9 was neuronal differentiation [1], later it was found that PCSK9 is involved in LDL cholesterol metabolism.
The best-characterized role of the mature and secreted form of PCSK9 (the ~60 kDa cleaved enzyme with the ~14-kDa prodomain associated to the catalytic domain) is targeting LDLRs for degradation in the liver. The catalytic subunit binds the epidermal growth factor-A (EGF-A) domain of the LDLR at the hepatocyte cell surface leading to LDLR internalization and degradation.
Once LDL cholesterol binds LDLR, it enters the cell through clathrin-coated vesicles. After internalization, the acidic pH of endosomes disrupts the association of LDL cholesterol from its receptor. LDL particles remain within the endosome while a recycling vesicle returns the LDLR to the cell surface. Endosomes containing LDL cholesterol fuse with lysomes where LDL is degraded and cholesterol esters are hydrolyzed. The free cholesterol is then distributed to other cellular compartments. At the hepatocyte cell surface, the catalytic domain of PCSK9 can also bind LDLR. The complex is the internalized via clathrin-coated vesicles. Within the endosome, the affinity of PCSK9 for the LDLR is enhanced due to the low pH, preventing the recycling of the receptor to the cell surface. The complex is then directed to the lysosome, where both components, LDLR and PCSK9, are degraded [10][3]. In addition, in vitro studies in hepatocytes suggest that PCSK9 might also enhance intracellular LDLR degradation prior to its secretion. When PCSK9 binds to LDLR within the Golgi complex, there is an increase in the traffic of LDLR bound to PCSK9 from the trans Golgi network to lysosomes for degradation, instead of directing the receptors to the cell surface [11]. It has been suggested that PCSK9 might also induced LDLR degradation by ubiquitination of the receptor [12].
In addition to binding to LDLRs in the liver, PCSK9 has other less characterized roles in different tissues. In the small intestine, PCSK9 might regulate the production of triglyceride-rich apolipoprotein B and might also regulate transintestinal fecal cholesterol excretion. Furthermore, PCSK9 has been suggested to regulate the expression of very-low-density lipoprotein receptors (VLDLRs) in adipose tissue and the ApoE receptor 2 receptor in the brain, by means of a similar mechanism to the LDLR. PCSK9 binds VLDLR and ApoE receptor 2, finally resulting in the lysosomal degradation of the receptors. By modulation of ApoE receptor 2 and related anti-apoptotic signaling pathways, PSCK9 might regulate neuronal apoptosis. Endocrine pancreatic cells also expressed PCSK9, but it is not known the role of PCSK9 in pancreatic cells [3]. PCSK9 also inhibits epithelial Na+ channel (ENaC)-mediated Na+ absorption by reducing ENaC surface expression, mainly by an increased proteasomal degradation. By reducing ENaC channel number, PCSK9 might modulate epithelial Na+ absorption, which is a major contributor to blood pressure control [13].
PCSK9 and hypercholesterolemia
High serum levels of LDL cholesterol are strongly associated with a higher risk of developing cardiovascular disease (CVD), which is the leading cause of death worldwide.
People with familial hypercholesterolemia present mutations that cause defects in hepatic cholesterol clearance, leading to increased levels of LDL cholesterol in plasma and, therefore, increased risk of CVD. Previously, mutations in LDL receptor and apolipoprotein B (an essential component of LDL particles which interacts with LDLR) were linked to familial hypercholesterolemia. The studies performed during the last years have identified the gene encoding PCSK9 as a third locus related to familial hypercholesterolemia. There is a link between PCSK9 function and LDL cholesterol serum levels [2]. Thus, gain of function mutations in PCSK9 cause an infrequent form of familial hypercholesterolemia, while loss of function mutations are associated with hypocholesterolemia and reduced risk of CVD.
There is a relationship between lowering LDL cholesterol and reduction in risk of CVD. Hence, one of the main strategies to prevent CVD is reducing LDL cholesterol serum levels. The most used treatment to reduce LDL levels is statins, which inhibits 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase, a key enzyme in the synthesis of cholesterol. However, these drugs have some adverse effects including myopathy, rhabdomyolysis and hemorrhagic stroke, and some patients do not respond properly to the treatment. PCSK9 has become a promising target to treat hypercholesterolemia when statins are not a proper option. Inhibition of PCSK9 avoids LDLR degradation resulting in an increase of LDLR at the hepatocyte cell surface, enhancing the uptake of LDL cholesterol and lowering LDL cholesterol circulating levels. Several monoclonal antibodies against PCSK9 are being already used in phase III clinical trials, and there is an active research in other strategies to inhibit PCSK9 (discussed below).
PCSK9/LDLR complex
The interaction between PCSK9 and LDL receptor can be explained by a two-step model. In the first step, the Pro-Cat domain of PCSK9 initiates contact with EGF-A of the LDL receptor at neutral pH. An is formed between residues 377– 379 of PCSK9 and residues 308–310 of EGF-A. The complex PCSK9:LDLR is internalized and exposure to the low pH environment of the endosome, where the CT domain of PCSK9 binds the Ligand-Binding domain of LDLR. This interaction impair the ability of the receptor to adopt a recycling-competent conformation and promote trafficking of the PCSK9-LDLR complex to the lysosome (Yamamoto, Lu et al. 2011).
In the absence of PCSK9, lipoprotein binding to the LDLR leads to receptor-mediated endocytosis. The low pH environment of the endosome induces a conformational change in the LDLR, resulting in discharge of bound lipoprotein ligand and interaction between the β-propeller segment and ligand-binding repeats 4 and 5. This event permits the segregation and separate trafficking of the LDLR to the cell surface and the lipoprotein ligand to the lysosome, respectively (Yamamoto, Lu et al. 2011).
It is believed that the transition from neutral pH at the cell surface to low pH in the endosomal compartment activates a “histidine switch” that promotes the mentioned intramolecular interaction between receptor domains. A critical aspect of this conformational change is that it promotes ligand release, thereby facilitating receptor recycling to the cell surface, where it is available for another round of endocytosis. PCSK9-mediated interference with this process causes the LDLR to traffic to lysosomes, where it is degraded (Yamamoto, Lu et al. 2011).
Kinetics of PCSK9
Under normal conditions, PCSK9 has a half-life in plasma of approximately 5 minutes. It has been showed that in humans and mice, LDLR is a major regulator for PCSK9 levels and clearance, therefore in the presence of an additional copy of LDLR in the liver (induced by transgenic expression) reduces the half-life of PCSK9 by 50%, to 2.9 minutes, whereas in the absence of LDLR, the half-life of PCSK9 in serum is prolonged between 3–10 times above normal.
The kinetics of wild-type (WT) PCSK9 binding to LDLR shows Kd(poner d pequeña) values that range from 90 to 840 nM at neutral pH, and its affinity to LDLR becomes ∼100-fold higher at lower pH with Kd(poner d pequeña) values ranging from 1–8 nM. (Relacionar con los cambios estructurales de arriba)
PCSK9 binding to LDLR has been described as biphasic, with a first rapid phase characterized by a half-time of 6.6 minutes, which accounts for 35% of the equilibrium binding and a second slow phase whose half-time is 94 minutes. Similarly, 25% of the PCSK9 bound to LDLR dissociates during the rapid phase with a half-time of 19 minutes, while the remaining PCSK9 dissociates slowly with a half-time of 297 minutes.
Despite the rapid binding of PCSK9 and internalization of LDLR by hepatocytes, PCSK9-mediated degradation of LDLR in vitro has only been observed after several hours. It was further shown that, at least in mice, PCSK9 remains intact in the liver for up to 4 hours after its internalization, thus suggesting that other events might be required in order to allow PCSK9-mediated degradation of LDLR (or LDLR mediated degradation of PCSK9).
(Giunzioni and Tavori 2015)
You may include any references to papers as in: the use of JSmol in Proteopedia [14] or to the article describing Jmol [15] to the rescue.
PCSK9 as a therapeutic target
Regarding the fact that PCSK9 reduces the LDL-R and thus LDL-C clearance from blood, high concentrations of this protein in plasma increase risk to suffer from cardiovascular diseases. Effect that has been considerably reversed when studying loss of function mutations in PCSK9 which conditioned self-processing and secretion. Hence, several studies approaching alternative treatments against hypercholesterolemia used PCSK9 as new target to avoid the side effects in statins treatment. To this end inhibitory strategies against PCSK9 are under investigation with different approaches that can either prevent the protease from binding LDL-R or inhibiting its synthesis and processing.
Extracellular inhibitors
Their strategy is focused in the reduction of PCSK9 function or its plasma level. The two main inhibitors are:
Monoclonal antibodies
They constitute the most successful strategy via sequestrating in plasma circulating PSCK9 binding to a specific epitope in the molecule. By
binding to the catalytic domain and prodomain of the protease, they neutralize PCSK9 activity, thus, preventing its interaction with LDL-R.
In clinical trials they reached a maximum a suppression of plasma free PCSK9 after 4 to 8 hours of administration, achieving a 65% reduction of
LDL-C in healthy patients and a 60 to 80% reduction in patients with hypercholesterolemia.
There are three known mAb that have reached the clinical trials evolocumab, alirocumab and bococizumab. Only the two first antibodies are fully human while the last one is approximately 3% murine which has been withdrawn due to anti-drug antibodies responses [3]. Out of metanalyses it has been addressed that they reduce cardiovascular mortality as well as the rate of myocardial infarction. Both alirocumab (Praluent®) and evolocumab (Repatha®) received FDA and EMA approval and are indicated as complement to diet and maximally tolerated therapy for the treatment of adults with heterozygous familial hypercholesterolemia or clinical atherosclerotic CVD requiring additional lowering of LDL-C [16].
Pharmacodynamics:
Antibodies interaction with PCSK9 is based in EGFA binding site of the peptidase. In affinity studies, unravelling of the mechanism of
interaction of antibodies with PCSK9 was carried out using an antibody phage library. Among them, the one which most potently inhibited
PCSK9/LDLR was antibody 33 (Fab33) also known as RG7652 causing a reduction of LDL-C levels in humans. Its is centered on EGFA binding
site and the antibody engages to it by 5 of its 6 complementary-determining region (CDR) loops (H1, H2, H3, L1, L3). As well, an additional hydrogen bond is formed by residues near the heavy chain residue 73. An approximated 950 Å2 surface area is buried at each side of Fab33-PCSK9 contact, with 73% of this area buried by the heavy chain. Thus, mechanism underlying the interaction is based in the CDR-H2 loop of the Fab33 which is projected to the N terminal groove of PCSK9 which is normally occupied by P' helix. Consequently P' helix, which by its P1' Ser 153 and P3' Pro 155 residues stabilize the bound of PCSK9 to LDLR-EGFA domain (via polar and Van der Waal interactions), is displaced and cleaved (In downstream P'helix Arg165-Tyr166 residues)[17].
Pharmacokinetics:
Recommended doses are 75 mg and 140 mg for alirocumab and evolocumab respectively via subcutaneous (SC) administration every 2 weeks. In case of
uptriation for an additional lowering effect entails 150 mg dose every 2 weeks for alirocumab and 420 mg dose every month for evolocumab, both
subcutaneously. Specifically, for alirocumab 75 mg SC administration in a Phase I study caused a complete loss of free PCSK9 between day 3
and 4 causing a maximal reduction of LCL-R at day 5. Compared to this, 150 mg reached the same effect in just one day and persisted for 10 days.
Uptriation dosage in alirocumab achieved a reduction below 70 mg/Dl in LDL-C in 79.3% of patients. Also this dosage adjustment for both ab
increased HDL-C by 4.6 % and 7 % in alirocumab and evolocumab as well as 2.9% and 4.2% increase of apoA1 lipoprotein respectively. Upon PCSK9
binding to the mAb, LDLR levels increased, thus, more LDL particles where internalized [16].
They are effective both as monotherapy or combined with statins at the maximum tolerable dose, besides this, they reduce lipoprotein A up to a 30% which is a risk factor for development of CVD. The average half-life of mAb is 2.5-3 days and the elimination of the complex with PCSK9 may probably have a similar mechanism to PCSK9-mediated degradation of LDLR via endosomal/lysosomal route [18].
PCSK9 binding Adnectins
They are a group of proteins based in the 10th type III domain of human fibronectin. Their affinity and specificity towards the
therapeutical target is increased by molecular engineering of its loops via introduction of surfaces that bind to it. Similarly to the variable
regions in the antibodies, they have β sheet fold structures with diversified loops. One of these molecules studied in clinical trials is known
as BMS-962476, a 11kDa polypeptide combined with polyethylene glycol (PEG) which increases its pharamacokinetics to subnanomolar affinity
binding.The molecule is apparently safe, well tolerated and rapidly reduces free PCSK9 (90% with > 0.3 mg/kg dose) and LDL-C, this last
achieving 48% maximal dose-related reduction. The duration of their effects is dose dependent the lower the dose the faster the return to base
line levels of PCSK9 [3].
Pharmacodynamics:
When bound, BMS-962476 progenitor adnectin covers 910 Å2 of PCSK9 surface close to the LDLR binding site. It binds to a concave pocket (compromising just 37 aminoacids in human PCSK9) in the catalytic domain generating contacts with the residues from N-terminus and FG loop of adnectin. The loop constitutes approximately the 70% of consisting in a chain of stacked residues together with PCSK9, while N-terminus contacts solely with D 374 residue (where one of the possible gain of function mutations can occur) of the catalytic domain.
FG loop conformation is stabilized thanks to interactions of β-sheet, hydrophobic and charge to charge nature with several protease residues. They might be binding in couples of two adnectins and two PCSK9 molecules. Due to the interaction, PCSK9 suffers a conformation change in which the loop of 212-218 residues in the asymmetric unit is partially folded away from catalytic site burying one of its residues in the prodomain of the second molecule in the asymmetric unit. Consequently, adnectin and EGFA cannot simultaneously bind to PCSK9 which is competent with BMS-962476 ability to competitively displace LDLR EGFA binding to PCSK9.
Kd value at 37º is of 1.3± 0.2 nM consequently they have an elevated binding affinity. Adnectin causes a potent inhibition of the PCSK9 with an IC50 of 2.0 ± 0.6 nM and in cell based inhibition assays the adnectin restored completely LDLR activity with an EC50 of 31 nM. Thus, preventing the binding and LDLR-PCSK9 cointernalization, increasing receptor recycling and LDL uptake. In transgenic mice expressing human PCSK9, BMS-962476 reduced potently free plasma PCSK9 with an ED50 of approximately 0.01mg/KG. For those mice overexpressing PCSK9 with a strong cholesterol phenotype, cholesterol fell approximately a 35%, 3 hours after intraperitoneal injection of BMS-962476 (levels returned to baseline after 48 hours). As well, level of human PCSK9 in plasma rapidly decreased to 0 due to adnectin high affinity and fast binding to circulating PCSK9. Consistent with this, there was a lowering in plasma apo B and apo E containing lipoproteins concentration and an upregulation of LDLR activity in the liver. The percentage of free protease was rapidly supressed in more than 99% together with the reduction of 55% of cholesterol in cynomolgus monkeys treated with BMS-962476.
Pharmacokinetics: The average half life of BMS-962476 is of 108h and has an elevated clearance with a Vd of 86 ml/kg after a 5mg/kg dose administered to cynomolgus monkeys. BMS-962476 had a 79% of subutaneous bioavalability thus is likely to be well absorbed in humans after subcutaneous administration. It is rapidly filtered by the kidney (requiring pharmacokinetics enhancement modification for in vivo applications) [19].
A recent study which is currently in phase I trial used as strategy to defeat PCSK9 activity a peptide based vaccine, AT04A, in atherogenic mouse model. It consists in a PCSK9 peptide conjugated to an immunogenic carrier protein which elicits T helper activity. Inducing high persistent levels of ab against PCSK9, a significant reduction of plasma total LDL-C (-53%) as well as a reduction in the atherosclerotic lesion area (- 60%) [16].
Intracellular inhibitors
Small interference RNA (siRNA)
In clinical trials it has been used the ALN-PCSsc RNA also known as inclisiran which is a long-acting iRNA taken up by hepatocytes. A 48% of patients in Phase II with high risk of CVD and high levels of LDL-C showed reduced levels of LDL-C below 50mg/dl, in a 2 dose-regime with 300 mg of this RNA [16].
Antisense oligonucleotides (ASOs)
The most know was SPC5001A 14-mer oligonucleotide that did not go further the Phase I trial in the clinical development due to the acute injection side reactions and the development after increasing the dose of an acute tubular neurosis [16].
CRISPR-Cas 9 gene edition
It has reduced plasma PCSK9 after inducing a non-sense mutation in hepatic tissue in mice with no evidence of off target mutagenesis in preclinical studies [16].
Small molecule therapeutics
Created with the approach of blocking PCSK9 secretion to serum, PF-06446846 is a compound able to interact with the ribosome exit site while PCSK9 is being synthesized generating a gridlock inhibiting the obtainment of the final product. Still, it is in preclinical stage and its study is discontinuous [20].
Future therapeutics
Based in the flexibility of the P' helix structure in charge of stabilizing the bound formed between PCSK9 and EGFA-LDLR domain, efficacy of hypercholesterolemia treatments can be increased. To achieve it, a recent approach against PCSK9 uses engineered small molecules, administered orally which target non-easily accessible regions close to the EGFA binding domain. P' helix is normally close to N-terminal groove which is next to the EGFA binding site where is possible to target small peptides conjugated to a peptide analogous to EGFA and selectively inhibit PCSK9 binding to LDLR. This is possible thanks to the weak interactions of P' helix with N-terminal groove, favouring the non-contact conformation or “out” state of this structure. Thus, they consist in Pep 2-8, a 13 aminoacids peptide homologous to EGFA to target its binding site in PCSK9, conjugated to a peptide extension which will firmly bind it to the protease in the N-terminal groove. The extension peptides used to generate the antagonism have the following coding MESFPGWNLV(hR)IGLLR and SFAFPGWNLV(hR)IGLLR.
Then, the specificity of the peptide extension to the N-terminal groove is based in the imitation of the helical structure and folding of the P´helix. Consequently, the extension needs to have:
- (I) A WNLxRI residues motif (being x any given aminoacid) with a helical conformation resembling P´helix.
- (II)The ability to fold back towards the EGFA binding site after this motif same as P' helix does when EGFA is bound to PCSK9 and so creating contacts with the analogue.
- (III) An helix capping hydrogen bond donor, then, a residue able to interact with the N-terminal groove residues and anchor firmly.
- (IV) A mechanism to tether the C-terminal tail onto the WNLxRI domain, helping the folding of the motif to the helical conformation, thus reducing the entropic cost of binding the groove.
The inhibitory activity is based in the introduction of modifications in the protein extension to reduce the favourable interactions of PCSK9 with the LDLR-EGFA. The mechanism is based the presence of certain aminoacids that enable the ability to extend toward the EGFA binding site. In this regard, antagonism is based in the steric clash of EGFA residues Leu 298, Asp299 and Asn 300 with the Pro 5 residue from the peptide. Furthermore, the presence of a common phenylalanine/tyrosine–proline–glycine (FPG/YPG) common domain in the extension peptide adopting a β-turn conformation also antagonize the binding of LDLR receptor[17].
Disease
Relevance
Structural highlights
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.

