HIV and accessory proteins
From Proteopedia
(Difference between revisions)
| Line 15: | Line 15: | ||
=== Viral infectivity factor (Vif) === | === Viral infectivity factor (Vif) === | ||
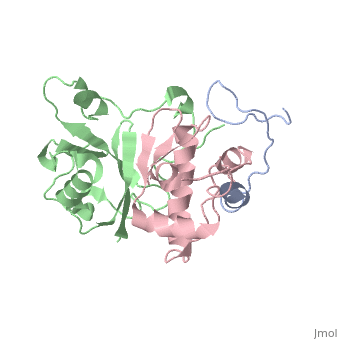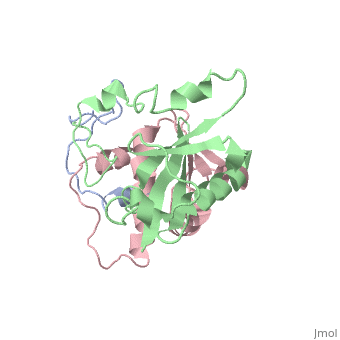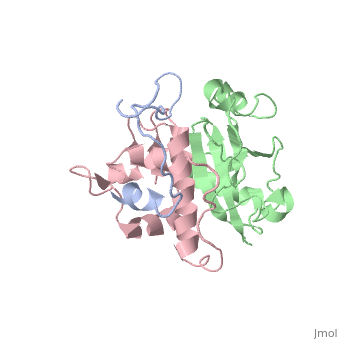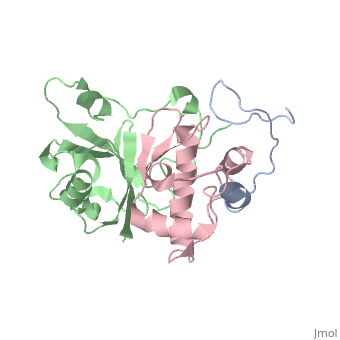
<scene name='71/719207/Cv/1'>Figure 1. Viral infectivity factor (Vif) protects the virus from host defenses mounted by APOBEC3G</scene> (Protein Data Bank ID: [[2ma9]]). | <scene name='71/719207/Cv/1'>Figure 1. Viral infectivity factor (Vif) protects the virus from host defenses mounted by APOBEC3G</scene> (Protein Data Bank ID: [[2ma9]]). | ||
| - | |||
| - | |||
Viral infectivity factor (Vif) is an accessory protein with a complex, cell-specific role in the pathway of HIV. In typically non-permissible cell types, the normal targets of HIV (lymphocytes, macrophages, and some types of T-cells), Vif interacts with the cell immunity mechanism factor APOBEC3G. APOBEC3G functions by becoming incorporated into the virions, and preventing reverse transcription from occurring. Vif counteracts this by binding to APOBEC3G and tagging it to be destroyed by polyubiquitination. Vif’s functionality makes it unnecessary in permissible cell types where APOBEC3G is not present. | Viral infectivity factor (Vif) is an accessory protein with a complex, cell-specific role in the pathway of HIV. In typically non-permissible cell types, the normal targets of HIV (lymphocytes, macrophages, and some types of T-cells), Vif interacts with the cell immunity mechanism factor APOBEC3G. APOBEC3G functions by becoming incorporated into the virions, and preventing reverse transcription from occurring. Vif counteracts this by binding to APOBEC3G and tagging it to be destroyed by polyubiquitination. Vif’s functionality makes it unnecessary in permissible cell types where APOBEC3G is not present. | ||
| - | |||
Vif is made up of at least two crucial domains, the N terminus of the protein which binds to APOBEC3G, and The C terminal region which contains many hydrophobic residues and is essential for the degradation of APOBEC3G. The complex workings of this protein led to the determination of its functionality taking a long time. The elucidation of this function could lead to the possible development of drugs to combat HIV <ref name="Rose" />. | Vif is made up of at least two crucial domains, the N terminus of the protein which binds to APOBEC3G, and The C terminal region which contains many hydrophobic residues and is essential for the degradation of APOBEC3G. The complex workings of this protein led to the determination of its functionality taking a long time. The elucidation of this function could lead to the possible development of drugs to combat HIV <ref name="Rose" />. | ||
| Line 29: | Line 26: | ||
Vpr can facilitate the movement of HIV cDNA from the cytoplasm to the nucleus where it can be utilized by the cell. Another function of Vpr is the ability to activate the transcription of HIV promoters. Vpr also possesses the ability to halt cell division between the growth 2 phase and mitosis. This activity is correlated with a decrease in the activity of p34cdc2, a crucial element for the progression from G2 to mitosis. After this halt in their growth cycle the affected cells undergo apoptosis <ref name="Yao">Yao, S., Torres, A.M., Azad, A.A., Macreadie, I.G., Norton, R.S. (1998) Solution structure of peptides from HIV-1 VPR protein that cause membrane permeabilization and growth arrest. [http://www.rcsb.org/pdb/explore/explore.do?structureId=1BDE J.Pebt.Sci. 4.426-435]</ref>. | Vpr can facilitate the movement of HIV cDNA from the cytoplasm to the nucleus where it can be utilized by the cell. Another function of Vpr is the ability to activate the transcription of HIV promoters. Vpr also possesses the ability to halt cell division between the growth 2 phase and mitosis. This activity is correlated with a decrease in the activity of p34cdc2, a crucial element for the progression from G2 to mitosis. After this halt in their growth cycle the affected cells undergo apoptosis <ref name="Yao">Yao, S., Torres, A.M., Azad, A.A., Macreadie, I.G., Norton, R.S. (1998) Solution structure of peptides from HIV-1 VPR protein that cause membrane permeabilization and growth arrest. [http://www.rcsb.org/pdb/explore/explore.do?structureId=1BDE J.Pebt.Sci. 4.426-435]</ref>. | ||
| - | |||
| - | |||
=== Viral protein U (Vpu) === | === Viral protein U (Vpu) === | ||
<scene name='71/719207/Cv/3'>Figure 3. Viral protein U is involved with the release of the virion</scene> (Protein Data Bank ID: [[1pi8]]). | <scene name='71/719207/Cv/3'>Figure 3. Viral protein U is involved with the release of the virion</scene> (Protein Data Bank ID: [[1pi8]]). | ||
| - | |||
Viral protein U (Vpu) is another accessory protein that functions in increasing the virulence of HIV-1. | Viral protein U (Vpu) is another accessory protein that functions in increasing the virulence of HIV-1. | ||
How Vpu works is not completely understood but it is believed to aid in the release of virions by causing the tethering proteins to release the virions on the outside of the cell. When Vpu is not present, the virions accumulate on the outside of the cell and are not released, this is why it is believed that they cause the release of virions from the infected cell. | How Vpu works is not completely understood but it is believed to aid in the release of virions by causing the tethering proteins to release the virions on the outside of the cell. When Vpu is not present, the virions accumulate on the outside of the cell and are not released, this is why it is believed that they cause the release of virions from the infected cell. | ||
| - | |||
| - | |||
| - | |||
===HIV-1 Protease (P6)=== | ===HIV-1 Protease (P6)=== | ||
| Line 50: | Line 41: | ||
Negative regulatory factor (Nef) performs two functions in the host cell. The first is that it is involved in T cell activation. The second is that it maintains a persistent state of infection. | Negative regulatory factor (Nef) performs two functions in the host cell. The first is that it is involved in T cell activation. The second is that it maintains a persistent state of infection. | ||
| - | |||
Nef downregulates the expression of surface proteins important to the host immune response, such as the major-histocompatibility complexes and CD4 and CD28 on helper T cells <ref name="Das1" />. | Nef downregulates the expression of surface proteins important to the host immune response, such as the major-histocompatibility complexes and CD4 and CD28 on helper T cells <ref name="Das1" />. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
=== Regulator of virion (Rev) === | === Regulator of virion (Rev) === | ||
<scene name='71/719207/Cv/5'>Figure 5. Regulator of virion (Rev) exports unspliced RNA transcripts out of the nucleus</scene> (Protein Data Bank: [[4pmi]]). | <scene name='71/719207/Cv/5'>Figure 5. Regulator of virion (Rev) exports unspliced RNA transcripts out of the nucleus</scene> (Protein Data Bank: [[4pmi]]). | ||
| - | |||
Regulator of virion (Rev) functions later in the pathway of HIV. It exports unspliced RNA out of the nucleus and into the cytoplasm. This unspliced RNA is thus ensured to be incorporated during assembly of the virion particle. | Regulator of virion (Rev) functions later in the pathway of HIV. It exports unspliced RNA out of the nucleus and into the cytoplasm. This unspliced RNA is thus ensured to be incorporated during assembly of the virion particle. | ||
| - | |||
Rev binds to viral RNA factor, Rev response element (RRE), available on most unspliced transcripts. Rev then escorts these unspliced transcripts out of the nucleus, utilizing the Crm1 pathway. Once in the cytoplasm, these transcripts either become the template for translation, or they are assembled into virion particles <ref name="Blissenbach" />. | Rev binds to viral RNA factor, Rev response element (RRE), available on most unspliced transcripts. Rev then escorts these unspliced transcripts out of the nucleus, utilizing the Crm1 pathway. Once in the cytoplasm, these transcripts either become the template for translation, or they are assembled into virion particles <ref name="Blissenbach" />. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
=== Trans-activator of transcription (Tat) === | === Trans-activator of transcription (Tat) === | ||
<scene name='71/719207/Cv/6'>Figure 6. Trans-activator of transcription (Tat) coupled with pTEFb, which stimulates transcription. Tat performs several other functions in the cell as well</scene> (Protein Data Bank ID: [[3mia]]). | <scene name='71/719207/Cv/6'>Figure 6. Trans-activator of transcription (Tat) coupled with pTEFb, which stimulates transcription. Tat performs several other functions in the cell as well</scene> (Protein Data Bank ID: [[3mia]]). | ||
| - | |||
| - | |||
One way HIV regulates transcription and gene expression is through the trans-activator of transcription (Tat). | One way HIV regulates transcription and gene expression is through the trans-activator of transcription (Tat). | ||
| - | |||
Tat acts early on in the HIV cycle. It functions to activate transcription by interacting with the long terminal repeat located in the trans-activator response (TAR) hairpin. Tat binding at this region promotes the binding of transcription factor, pTEFb, which can affect RNA polymerase II processivity and formation of transcription complexes. Tat has many other proposed functions, including recruitment of proteins involved in chromatin-remodeling, gene regulation, RNA processing, translation, and reverse transcription <ref name="Das2" />. | Tat acts early on in the HIV cycle. It functions to activate transcription by interacting with the long terminal repeat located in the trans-activator response (TAR) hairpin. Tat binding at this region promotes the binding of transcription factor, pTEFb, which can affect RNA polymerase II processivity and formation of transcription complexes. Tat has many other proposed functions, including recruitment of proteins involved in chromatin-remodeling, gene regulation, RNA processing, translation, and reverse transcription <ref name="Das2" />. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
== Pathway == | == Pathway == | ||
During early infection, HIV is macrophage tropic, meaning that it only infects natural killer cells, CD8+ killer T cells, macrophages, cells of the nervous system, and dendritic cells. During that phase, the cells harbor, replicate, and bud HIV without lysis. On the contrary, during later infections, HIV is T-lymphocyte tropic, meaning it infects CD4+ T lymphocytes and will lyse them during production of new viruses, making it a productive infection. HIV enters a cell by interacting its gp120 portion, the head portion of the protruding glycoprotein, with CD4, the major cellular receptor that is abundantly present on T lymphocytes. This interaction causes a conformational change in gp120 and gp41, the spike portion of the protruding glycoprotein, resulting in the exposure of new binding regions on these proteins, thus causing a co-receptor reaction with either CCR5, the macrophages’ co-receptor, or fusin, the T lymphocyte co-receptor. The virus then embeds into the membrane of the CD4+ cell. The gp41 protein changes into a coiled shape that brings the virus and cell membrane close to each other, allowing the fusion of the virus’ membrane and the host cell, resulting in entry and uncoating, which is the release of viral nucleic acids. In the cytoplasm, the viral RNA is converted to DNA by the HIV-encoded reverse transcriptase. The reverse transcriptase binds to the tRNAlys, which is bound to the RNA genome that serves as a primer for the reverse transcriptase, initiating DNA synthesis. When the reverse transcription is completed, it produces a double-stranded DNA (dsDNA) that contains long terminal repeats (LTRs) located at the end of the viral genome. The viral DNA is transported to the nucleus where it may be integrated into the host’s chromosomal DNA, which is catalyzed by integrase protein found on the HIV <ref name="Shors">Shors, Teri. (2011). Understanding Viruses. (2nd ed., pp. 507-511). Oshkosh, Wisconsin: Jones and Bartlett.</ref>. | During early infection, HIV is macrophage tropic, meaning that it only infects natural killer cells, CD8+ killer T cells, macrophages, cells of the nervous system, and dendritic cells. During that phase, the cells harbor, replicate, and bud HIV without lysis. On the contrary, during later infections, HIV is T-lymphocyte tropic, meaning it infects CD4+ T lymphocytes and will lyse them during production of new viruses, making it a productive infection. HIV enters a cell by interacting its gp120 portion, the head portion of the protruding glycoprotein, with CD4, the major cellular receptor that is abundantly present on T lymphocytes. This interaction causes a conformational change in gp120 and gp41, the spike portion of the protruding glycoprotein, resulting in the exposure of new binding regions on these proteins, thus causing a co-receptor reaction with either CCR5, the macrophages’ co-receptor, or fusin, the T lymphocyte co-receptor. The virus then embeds into the membrane of the CD4+ cell. The gp41 protein changes into a coiled shape that brings the virus and cell membrane close to each other, allowing the fusion of the virus’ membrane and the host cell, resulting in entry and uncoating, which is the release of viral nucleic acids. In the cytoplasm, the viral RNA is converted to DNA by the HIV-encoded reverse transcriptase. The reverse transcriptase binds to the tRNAlys, which is bound to the RNA genome that serves as a primer for the reverse transcriptase, initiating DNA synthesis. When the reverse transcription is completed, it produces a double-stranded DNA (dsDNA) that contains long terminal repeats (LTRs) located at the end of the viral genome. The viral DNA is transported to the nucleus where it may be integrated into the host’s chromosomal DNA, which is catalyzed by integrase protein found on the HIV <ref name="Shors">Shors, Teri. (2011). Understanding Viruses. (2nd ed., pp. 507-511). Oshkosh, Wisconsin: Jones and Bartlett.</ref>. | ||
During a productive infection, HIV-1 is reactivated and the lytic cell cycle occurs. The host’s transcriptional and translational machinery is used. The proviral DNA is transcribed by the cellular RNA polymerase II from a single promoter in the 5’ LTR into a 9 kb viral RNA primary transcript. Trans-activator of transcription (Tat) and regulator of virion (Rev) both play a role in the regulation of viral gene expression. Tat interacts with TAR, which is located at the 5’ end of all HIV RNAs, and drastically increases the transcription rate from the HIV LTR <ref name="Shors" />. Rev binds to the Rev response element (RRE), which is a well-conserved, 350 nucleotide element that’s located in the env coding region of the viral genome <ref name="Fernandes">Fernandes, J., Jayaraman, B., & Frankel, A. (2012). The HIV-1 Rev response element: An RNA scaffold that directs the cooperative assembly of a homo-oligomeric ribonucleoprotein complex.[http://doi.org/10.4161/rna.9.1.18178 RNA Biology, 9(1), 6–11.]</ref>. This binding facilitates the export of unspliced and incompletely spliced viral RNAs from the nucleus to the cytoplasm. Once they are in the cytoplasm, some of the full-length viral messenger RNAs are packaged into virions, whereas others are spliced within the nucleus to form mRNAs that are translated by the host’s ribosome into different viral proteins. The gag gene and a combination of both the gag and pol genes together are translated into large precursor polyproteins that are cleaved by a virus encoded protease. The env protein, gp160, is transported through the trans Golgi apparatus where it is glycosylated and cleaved by a cellular protease into gp120 and gp41 to form mature envelope proteins that are targeted to the surface of the infected cell. The gp120 and gp41 are held together by covalent bonds. Tat and other smaller HIV proteins overlap with the structural genes but are in different open reading frames. Their mRNAs are made by alternative splicing of the structural gene mRNAs and are translated. The viral RNA, gag, pol, and env proteins, the virion components of HIV, are assembled at budding sites located at the cellular membrane. Two copies of the full viral genomes that contain the primer tRNA are packed into virus particles <ref name="Shors" />. | During a productive infection, HIV-1 is reactivated and the lytic cell cycle occurs. The host’s transcriptional and translational machinery is used. The proviral DNA is transcribed by the cellular RNA polymerase II from a single promoter in the 5’ LTR into a 9 kb viral RNA primary transcript. Trans-activator of transcription (Tat) and regulator of virion (Rev) both play a role in the regulation of viral gene expression. Tat interacts with TAR, which is located at the 5’ end of all HIV RNAs, and drastically increases the transcription rate from the HIV LTR <ref name="Shors" />. Rev binds to the Rev response element (RRE), which is a well-conserved, 350 nucleotide element that’s located in the env coding region of the viral genome <ref name="Fernandes">Fernandes, J., Jayaraman, B., & Frankel, A. (2012). The HIV-1 Rev response element: An RNA scaffold that directs the cooperative assembly of a homo-oligomeric ribonucleoprotein complex.[http://doi.org/10.4161/rna.9.1.18178 RNA Biology, 9(1), 6–11.]</ref>. This binding facilitates the export of unspliced and incompletely spliced viral RNAs from the nucleus to the cytoplasm. Once they are in the cytoplasm, some of the full-length viral messenger RNAs are packaged into virions, whereas others are spliced within the nucleus to form mRNAs that are translated by the host’s ribosome into different viral proteins. The gag gene and a combination of both the gag and pol genes together are translated into large precursor polyproteins that are cleaved by a virus encoded protease. The env protein, gp160, is transported through the trans Golgi apparatus where it is glycosylated and cleaved by a cellular protease into gp120 and gp41 to form mature envelope proteins that are targeted to the surface of the infected cell. The gp120 and gp41 are held together by covalent bonds. Tat and other smaller HIV proteins overlap with the structural genes but are in different open reading frames. Their mRNAs are made by alternative splicing of the structural gene mRNAs and are translated. The viral RNA, gag, pol, and env proteins, the virion components of HIV, are assembled at budding sites located at the cellular membrane. Two copies of the full viral genomes that contain the primer tRNA are packed into virus particles <ref name="Shors" />. | ||
| - | + | ||
| + | The depletion of CD4+ helper T lymphocytes caused by HIV results in AIDS and a weakening of the immune system, permitting opportunistic infections to occur. The viral env proteins found on the surface of infected cells bind to uninfected cells causing the fusion of the plasma membrane, ultimately resulting in syncytia <ref name="Shors" />. | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references /> | <references /> | ||
Current revision
| |||||||||||
References
- ↑ The HIV Life Cycle. (2015) Retrieved from AIDSinfo
- ↑ Hemelaar, J. (2012). The Origin and Diversity of the HIV-1 pandemic.In Trends in Molecular Medicine, 18(3):182-192 DOI:10.1016
- ↑ Global HIV and AIDS Statistics. (2015). Retrieved from Averting HIV and AIDS.
- ↑ 4.0 4.1 Origin of HIV & AIDS. (2015). Retrieved from Averting HIV and AIDS.
- ↑ Where Did HIV Come From? (2011). Retrieved from The AIDS Institute
- ↑ 6.0 6.1 Rose, Kristine M., Marin, Mariana, Kozak, Susan L., Kabat, David. (2004). The viral infectivity factor (Vif) of HIV-1 unveiled. TRENDS in Molecular Medicine, 10(6), 291-297.
- ↑ Morellet, N., Bouaziz, S., Petitjean, P., Roques, B. P. (2003). NMR Structure of the HIV-1 Regulatory Protein VPR. J. Mol. Biol, 327, 215-227.
- ↑ 8.0 8.1 Oie Solbak, Sara Marie, Reksten, Tove Ragna, Hahn, Friedrich, Wray, Victor, Henklein, Petra, Henklein, Peter, Halskau, Oyvind, Schubert, Ulrich, Fossen, Torgils. (2013). HIV-1 p6 - a structured to flexible multifunctional membrane-interacting protein. Biochimica et Biophysica Acta, 1828, 816-823.
- ↑ 9.0 9.1 Das, S.R., Jameel, S. (2005). Biology of the HIV Nef protein. Indian Journal of Medical Research, 121(4), 315-332.
- ↑ 10.0 10.1 Blissenbach, M., Grewe, B., Hoffman, B., Brandt, S., Uberla, K. (2010). Nuclear RNA export and packaging functions of HIV-1 Rev revisited. Journal of Virology, 84(13), 6598-604.
- ↑ 11.0 11.1 Das, A. T., Harwig, A., & Berkhout, B. (2011). The HIV-1 Tat Protein Has a Versatile Role in Activating Viral Transcription. Journal of Virology, 85(18), 9506–9516.
- ↑ Yao, S., Torres, A.M., Azad, A.A., Macreadie, I.G., Norton, R.S. (1998) Solution structure of peptides from HIV-1 VPR protein that cause membrane permeabilization and growth arrest. J.Pebt.Sci. 4.426-435
- ↑ Demirov, Dimiter G., Orenstein, Jan M., and Freed, Eric O., (2002). The late domain of Human Immunodeficiency Virus type 1 promotes virus release in a cell type-dependent manner. Journal of Virology. 71(1), 105-117.
- ↑ 14.0 14.1 14.2 14.3 Shors, Teri. (2011). Understanding Viruses. (2nd ed., pp. 507-511). Oshkosh, Wisconsin: Jones and Bartlett.
- ↑ Fernandes, J., Jayaraman, B., & Frankel, A. (2012). The HIV-1 Rev response element: An RNA scaffold that directs the cooperative assembly of a homo-oligomeric ribonucleoprotein complex.RNA Biology, 9(1), 6–11.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Erin Bolger, Alexander Berchansky, Joel L. Sussman