We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Spindlin
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
== Function == | == Function == | ||
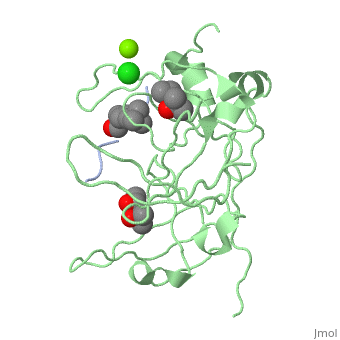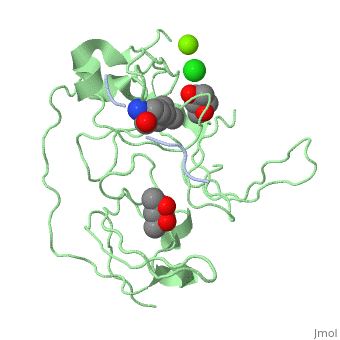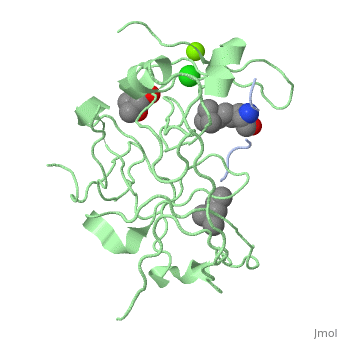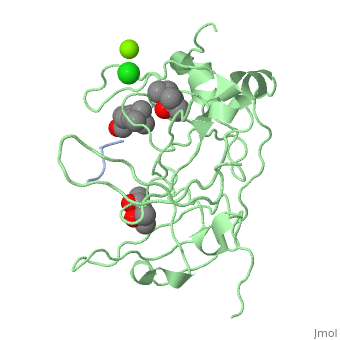
| - | '''Spindlin-1''' (SPIN1) is a Tudor-like domain-containing protein is a histone methylation effector protein and facilitates the expression of rRNA genes<ref>PMID:21960006</ref>. SPIN1 senses a cis-tail histone H3 methylation pattern | + | '''Spindlin-1''' (SPIN1) is a Tudor-like domain-containing protein is a histone methylation effector protein and facilitates the expression of rRNA genes<ref>PMID:21960006</ref>. SPIN1 senses a cis-tail histone H3 methylation pattern. '''Spindlin-2B''' is involved in the regulation of cell cycle progression. |
== Relevance == | == Relevance == | ||
Revision as of 09:15, 15 January 2018
| |||||||||||
3D Structures of spindlin
Updated on 15-January-2018
References
- ↑ Wang W, Chen Z, Mao Z, Zhang H, Ding X, Chen S, Zhang X, Xu R, Zhu B. Nucleolar protein Spindlin1 recognizes H3K4 methylation and stimulates the expression of rRNA genes. EMBO Rep. 2011 Oct 28;12(11):1160-6. doi: 10.1038/embor.2011.184. PMID:21960006 doi:http://dx.doi.org/10.1038/embor.2011.184
- ↑ Franz H, Greschik H, Willmann D, Ozretic L, Jilg CA, Wardelmann E, Jung M, Buettner R, Schule R. The histone code reader SPIN1 controls RET signaling in liposarcoma. Oncotarget. 2015 Mar 10;6(7):4773-89. doi: 10.18632/oncotarget.3000. PMID:25749382 doi:http://dx.doi.org/10.18632/oncotarget.3000
- ↑ Su X, Zhu G, Ding X, Lee SY, Dou Y, Zhu B, Wu W, Li H. Molecular basis underlying histone H3 lysine-arginine methylation pattern readout by Spin/Ssty repeats of Spindlin1. Genes Dev. 2014 Mar 15;28(6):622-36. doi: 10.1101/gad.233239.113. Epub 2014 Mar, 3. PMID:24589551 doi:http://dx.doi.org/10.1101/gad.233239.113