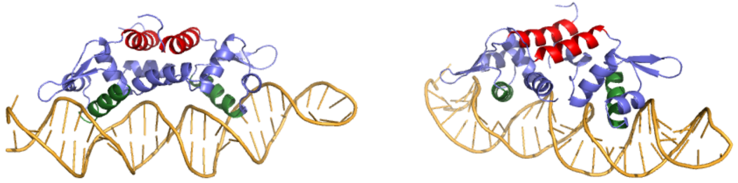We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1053
From Proteopedia
(Difference between revisions)
| Line 16: | Line 16: | ||
[[Image:800px-2KJB + 2KJC side by side.fw.png CROPPED.fw.png|600px|center|thumb| Figure 2: Comparison of Czr A bound to DNA to Czr A with Zn<sup>+2</sup> bound. α5 helices are shown in red and the α4 helices shown in green.]] | [[Image:800px-2KJB + 2KJC side by side.fw.png CROPPED.fw.png|600px|center|thumb| Figure 2: Comparison of Czr A bound to DNA to Czr A with Zn<sup>+2</sup> bound. α5 helices are shown in red and the α4 helices shown in green.]] | ||
== DNA Binding Site== | == DNA Binding Site== | ||
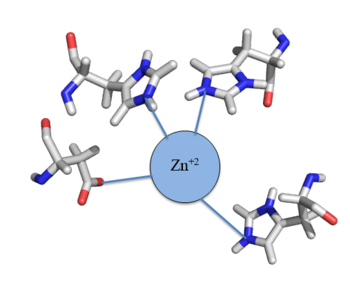
| - | Ser54, Ser57, and His58 are the primary residues involved in <scene name='69/694220/2kjb_colored/3'>DNA interaction</scene> with Czr A <ref name="critical"/>. These residues are likely to interact with the 5'-TGAA sequence found in the half-site of the DNA, where the α4 helices (green) <scene name='69/694219/Czra_with_dna/2'>form an interaction with DNA</scene> (Figure 3). Binding of two Zn<sup>+2</sup> ions <scene name='69/694220/Dna_residues_when_inhibited/2'>pushes these residues out of their DNA binding conformation</scene>. Additionally, Val42 and Gln53 (lime green) are involved in the <scene name='69/694220/Val_42_and_gln_53/1'>DNA binding pocket</scene>. | + | Ser54, Ser57, and His58 are the primary residues involved in <scene name='69/694220/2kjb_colored/3'>DNA interaction</scene> with Czr A <ref name="critical"/>. These residues are likely to interact with the 5'-TGAA sequence found in the half-site of the DNA, where the α4 helices (green) <scene name='69/694219/Czra_with_dna/2'>form an interaction with DNA</scene> (Figure 3). Binding of two Zn<sup>+2</sup> ions <scene name='69/694220/Dna_residues_when_inhibited/2'>pushes these residues out of their DNA binding conformation</scene>. Additionally, Val42 and Gln53 (lime green) are involved in the <scene name='69/694220/Val_42_and_gln_53/1'>DNA binding pocket</scene>. |
| + | |||
| + | The DNA bound state of Czr A was tested by using the known critical residues for DNA interactions . <scene name='69/694220/Dna_binding_residues/2'>Critical DNA binding residues</scene> | ||
| + | |||
| + | The Gln53 and Val42 (aqua) as well as the Ser54, Ser57, and His58 (lime) residues have been individually mutated to Ala, and DNA binding experiments were performed<ref name="critical"/>. Compared to wild type Czr A, mutating Gln53 and V42 residues resulted in an 11-fold and 160-fold decrease in K<sub>a</sub>, respectively. Mutations to the main DNA interaction sites Ser 54, Ser 57, and His 58 result in binding similar to the inhibited non-DNA binding state, suggesting that these residues are essential to binding DNA. While the conformational change that occurs from the Zinc to DNA bound state of Czr A is small,the alpha 4 helices (shown in green in Figure 2) are slightly shifted. The loss of DNA binding in the mutagenesis experiements in combination with the lack of any other major physical changes between these two states further suggests that the alpha 4 helices are the location of DNA binding in Czr A. Experimental data can be found in table 1 from this same article. | ||
[[Image:800px-DNABound Final.fw.png CROPPED.fw.png|750px|thumb|center| Figure 3: Two views of Czr A bound to DNA. A segment of DNA is shown in orange with the alpha 5 helices displayed in red and the alpha 4 helices shown in green]] | [[Image:800px-DNABound Final.fw.png CROPPED.fw.png|750px|thumb|center| Figure 3: Two views of Czr A bound to DNA. A segment of DNA is shown in orange with the alpha 5 helices displayed in red and the alpha 4 helices shown in green]] | ||
Revision as of 00:56, 19 January 2018
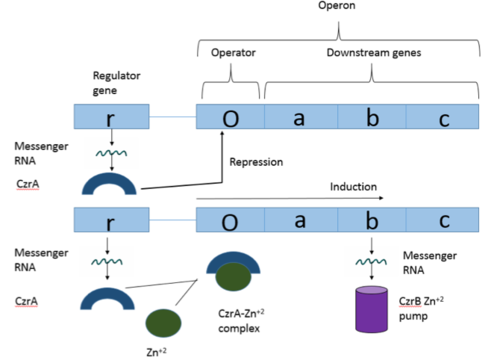
Zinc Dependent Transcriptional Repressor of the Czr operon (CzrA)
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Arunkumar A., Campanello G., Giedroc D. (2009). Solution Structure of a paradigm ArsR family zinc sensor in the DNA-bound state. PNAS 106:43 18177-18182.
- ↑ MacPherson S, Larochelle M, Turcotte B. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol Rev. 2006 Sep;70(3):583-604. PMID:16959962 doi:http://dx.doi.org/10.1128/MMBR.00015-06
- ↑ Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun 4;4(6):1609-1614.
- ↑ Grossoehme NE, Giedroc DP. Energetics of allosteric negative coupling in the zinc sensor S. aureus CzrA. J Am Chem Soc. 2009 Dec 16;131(49):17860-70. doi: 10.1021/ja906131b. PMID:19995076 doi:http://dx.doi.org/10.1021/ja906131b