Introduction
Ribonuclease A has been the subject of Nobel Prizes on Protein Folding and Solid Phase Peptide Synthesis.[1] The observation of ribonuclease folding helped Christian Anfinsen win the Nobel Prize in 1972 for his work on protein folding [2]. The presence of four disulfide bonds and two cis proline residues in the structure of RNase A greatly affects the structure and folding kinetics of RNase A [3]. When RNase A undergoes reductive denaturation, it spontaneously folds back on itself to form the same structure. The development of solid phase synthesis by Bruce Merrifield (Nobel Prize 1984) was a radical departure from traditional methods of bio-molecular synthesis that greatly increased efficiency. His method made possible the syntheses of much larger and more complex molecules; however, solid phase synthesis was not fully embraced until he demonstrated its full ability with the complete synthesis of Ribonuclease A.[1]
Protein Folding
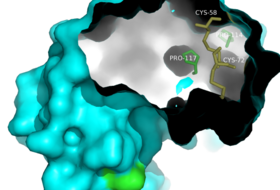
Residues important to the proper folding of RNase A. Locations of internal residues Pro-114, Pro-117, Cys-58, and Cys-72 are highlighted and labeled.
Interatomic interactions, delegated by the amino acid sequence, are responsible for formation of a protein's 3D structure [2]. Several of these interactions have been identified by the use of site directed mutagenesis to wildtype RNase A and subsequent comparison of the crystal structure to the wildtype. Although RNase A has 105 possible disulfide bond pairings, only one set of four bonds occurs. This unique observation leads to the "thermodynamic hypothesis", that a protein's native state is determined by the thermodynamic favorability of the whole system; thus the tertiary structure must be predetermined by intramolecular interactions within the amino acid sequence.[4] Since thermodynamic stability of a protein is affected by the environment's temperature, pH, and ionic strength, among other factors, the protein structure can only exist under physiological conditions. Today, the correlation between the amino acid sequence and the tertiary structure of RNase A continues to serve as a model for protein folding. Among the most important attributes of this model are noncovalent interactions (e.g. ), proline conformation, and disulfide bonding [5].
Proline Conformation
The presence of cis [3]proline residues plays a large role in protein folding. In nature, most amino acids reside in a trans conformation, but due to their cyclic structure, prolines are more stable in the cis conformation than any other amino acid. RNase A contains four proline residues, two reside in the cis conformation and two in the trans conformation.[5] The importance of these conformations are demonstrated based on the structure of RNase A variants with several mutations to the wild type amino acid sequence.
Located in an outer of RNase A, the peptide group of RNase A in its native state is found in the cis conformation. When proline was mutated to alanine, , a cis conformation still forms at position 93 which is an energetically unfavorable conformation for an alanine residue [6]. Upon unfolding, Tyr92-Ala93 undergoes isomerization to form its more favorable trans conformation demonstrating that the cis conformation is favored by other interactions within the folded protein structure. Although the overall structure of RNase A is not affected by this mutation, the rate of folding greatly decreases upon insertion of the P93A mutation, suggesting an important kinetic contribution of cis prolines to protein folding.[6]
The peptide bond also resides in a cis conformation in its folded structure, but exists in the trans conformation in its unfolded state; therefore, steric restraints imposed by the rest of the protein must be responsible for this cis conformation. Unlike P93A, the insertion of a point mutation causes the peptide bond to adopt a trans conformation and causes a 9.3 Å movement of the loop [7]. The kinetic rate and overall native conformation are not significantly effected by this mutation; however, locally, a rearrangement of the hydrogen-bonding network occurs. Results of this mutation confirm that steric hinderance of the protein can lead to formation of the cis conformation by a proline and is further energetically stabilized by hydrogen bonding, Van der Waals, and electrostatic interactions within the protein.
Another important role of proline residues is their involvement in β turns. β turns are 180° turns commonly found in globular proteins to allow for a compact structure by connecting the ends of adjacent antiparallel β sheets [4]. The turn consists of a sequence of four amino acid residues. The carbonyl of the first amino acid hydrogen bonds with the amino group of the fourth amino acid. Proline is involved in β turns because it is small, flexible, and assumes a cis conformation, all attributes that allow for formation of a turn. In RNase A both Pro93 and Pro114 are involved in β turns.[1] Proline residues are important to protein folding because their ability to form a favorable cis conformation allows for thermodynamic favorability of β turn formation. With β turns, amino acids can fold back on themselves allowing the protein to reside in a compact, globular structure.
Disulfide Bonds
Another important feature of the folding of RNase A is the presence of four disulfide bonds. These bonds contribute to the thermal stability and the rate of folding of RNase A. The residues involved in these linkages include , , , and . Cys26-Cys84 and Cys58-Cys110 stabilize an interaction between an α-helix and a β-sheet which is the main contributor to the thermodynamic stability of the enzyme.
Measurements of protein activity upon removal of disulfide bridges show that the change in enzymatic activity is very small and that not all disulfide bridges are essential for the structure or the reactivity of the protein. However, removal of disulfide bonds does destabilize the hydrophobic core and decreases the rate of folding. RNase A actually has a rate-determining three-disulfide intermediate. An analog of this, , shows RNase A, missing the disulfide bond, Cys40-Cys95, that would normally occur here. In the variant, only 3 disulfide bonds are present, but the overall structure is only changed slightly. The differences occur in residues in close proximity to the location of the missing disulfide bond, and , where there are increased levels of disorder and a destabilized hydrophobic core [6].
Medical Importance of Protein Folding
Protein folding has several medical implications. Diseases such as ALS, Alzheimer's Disease, and Parkinson's Disease can all be traced back to protein folding because proteins can form aberrant aggregates when they do not fold correctly. This abnormality can be toxic to human nerve cells. All proteins contain . The hydrophilic residues lie on the outer part of the protein and the hydrophobic residues bury themselves within the interior of the protein due to the hydrophobic effect [5]. Mistakes made during protein folding may cause a protein to expose and, in turn, cause several proteins to stick together and form a plaque. In the future researchers hope to design drugs that combat mistakes in protein folding [8]. The use of ribonuclease A in protein folding research has been an instrumental feature in designing experiments to determine these "misfolding" snapshots and in developing therapies to prevent protein misfolding.
Summary of Protein Folding
Protein folding is not due to one interaction, but a network of interactions within the protein. When a proline residue or disulfide bond is removed from RNase A, the structural changes are usually confined to the site of mutation and minor structural changes occur within close proximity to the mutation. Although the effects of mutations seem to be localized, mutating proteins greatly effects the stability of the molecule and the rate of folding.
Semisynthetic Ribonuclease A
Peptide Synthesis
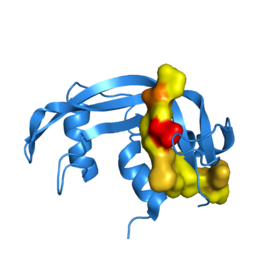
Semisynthetic RNase A. The synthetic peptide analog, RNase 111-118, is colored according to hydrophilicity. Yellow areas are comprised of hydrophobic residues. Red and brown segments are negatively and positively charged residues, respectively.
The peptide synthesis of non-natural and non-coded proteins allowed scientists to analyze the mechanism and structure-activity relationships of classical enzyme molecules that were not accessible by traditional biomedical methods. These syntheses, though, were both difficult and time consuming, and advances in technique developed slowly[9]. At the beginning of the twentieth century, Emil Fischer performed the first synthesis of a peptide, but it was not until 1953 that the first peptide hormone was synthesized by Du Vigneaud[9]. The development of solid phase synthesis by Bruce Merrifield was a radical departure from traditional methods of bio-molecular synthesis that greatly increased efficiency. His method made possible the syntheses of much larger and more complex molecules; however, solid phase synthesis was not fully embraced until he demonstrated its full ability with the . This milestone synthesis and subsequent semisynthetic syntheses of enzymes including enriched the hypothesis that the amino acid sequence of a protein contains all necessary information to direct the formation of a fully active enzyme and, additionally, that an enzyme demonstrating the catalytic capacity and specificity of a naturally produced enzyme can be made in laboratory[9][10][11].
Peptide synthesis is the production of proteins in which multiple amino acids are linked together through peptide bonds. A general chemical requirement for peptide synthesis is the blockage of the carboxyl group of one amino acid and the amino group of the second amino acid. The carboxyl group of the free carboxyl group can be activated and the new peptide bond is formed[9]. A common type of peptide synthesis is the solid-phase synthesis, in which the end of the peptide chain is attached to a solid support.
Structure equals Function
Semisynthetic RNase A
The synthesis of semisynthetic RNasa A clearly exhibits the structure to function relationship that defines proteins. In the RNase A protein, the removal of six C terminal residues, leaving , completely halts enzymatic activity.[10] However, a complex of RNase 1-118 with a synthetic polypeptide comprising the C terminal residues restores enzymatic activity to RNase A. Upon the addition of the synthetic chain, the adopts a structure that closely resembles that of [10]. The restoration of the structure reconstitutes the enzymatic activity of RNase to 98%[10].
Also, the surface representation of semisynthetic RNase A illustrates the interface between the synthetic analog and the natural enzyme, 1srn.
The semi-synthetic RNase A comprises of residues 1-118 and the synthetic analog of residues 111-124. The RNase 1-118 was prepared by successive digestion of RNase A pepsin and carboxypeptidase A[12]. The synthetic component, RNase 111-124, was prepared by the use of solid-phase peptide synthetic methods, in which the peptide chain was assembled in the stepwise manner while it was attached at one end to a solid support. The peptide chain was extended by repetitive steps of de-protection, neutralization and coupling until the desired sequence was obtained[13]. It was important that the synthesis proceeds rapidly and in high yields to prevent side reactions or by-products.
Fully Synthetic RNase A
The demonstrates similar structural and functional characteristics (such as catalytic activity) as those of the RNase A[11]. The crystal data, X-ray data collection and refinement statistics show that the fully synthetic protein shares identical molecular structures with the wild type RNase A, and that the active sites of both emzymes contain no walter molecules and have no substrate ligand[11]. The crystal structure similarities of the , , and are further evidence that amino acid sequence dictates folded structure formation.
The peptide ligation chemistry in addition to solid-phase peptide synthesis is used to synthesize relatively longer peptide molecules with typical length of 125 residues[11]. The ligation methods overcome the length limitation of solid-phase synthesis, because the chemical ligation involves the joining of mutually reactive peptide segments created by solid-phase synthesis. The peptide bond in ligation is formed between an unprotected peptide and a peptide-thioester[11]. The shorter peptide segments are more rapidly prepared and are less susceptible to solubility issues in longer peptide chains.
The (124 residues) is prepared by two consecutive sets of one-pot ligations and related chemical transformations of six peptide segments (residues , , , , , , as highlighted in red)[11],which can prevent undesired byproduct formation. The six unprotected peptide segments were synthesized by highly optimized, stepwise solid-phase synthesis. This synthetic pathway is simple, has high overall yields, and it eliminate the need for the isolation of intermediate products.
Additional Proteopedia Pages about RNase A


