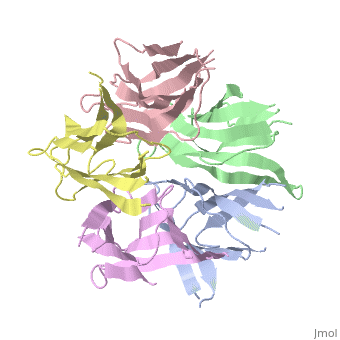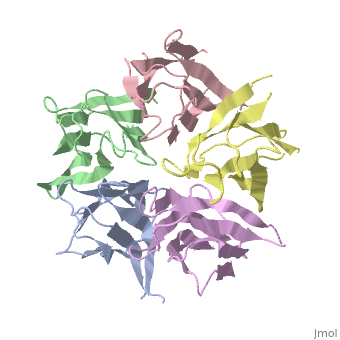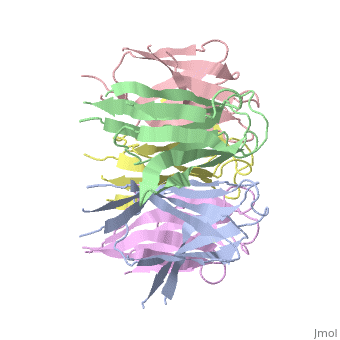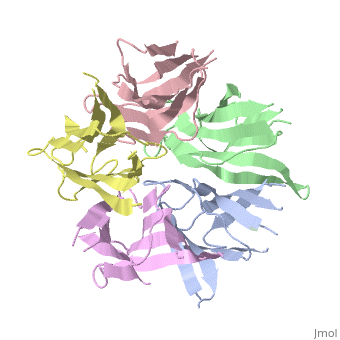This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Nucleoplasmin
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | |||
<StructureSection load='1k5j' size='350' side='right' caption='Nucleoplasmin core pentamer (PDB entry [[1k5j]])' scene=''> | <StructureSection load='1k5j' size='350' side='right' caption='Nucleoplasmin core pentamer (PDB entry [[1k5j]])' scene=''> | ||
| - | + | = Function = | |
'''Nucleoplasmin''' (Npl) is a molecular chaperone. Npl functions in the proper assembly of nucleosomes and chromatin structures<ref>PMID:15284896</ref>. | '''Nucleoplasmin''' (Npl) is a molecular chaperone. Npl functions in the proper assembly of nucleosomes and chromatin structures<ref>PMID:15284896</ref>. | ||
| + | He is participates in histone storage, sperm chromatin decondensation, and nucleosome assembly,enome stability, ribosome biogenesis, DNA duplication and transcriptional regulation. | ||
| + | In the assembly of nucleosomal arrays the NP transfer to DNA to him by binding the histones, this reaction requires ATP. | ||
| - | + | = Relevance = | |
Apoptotic chromatin condensation, one of the hallmarks of apoptosis, is regulated by Npl. | Apoptotic chromatin condensation, one of the hallmarks of apoptosis, is regulated by Npl. | ||
| - | </ | + | |
| + | =Structural highlights= | ||
| + | Nucleoplasmin (NP) is made out of five <scene name='46/467273/Np_monomer/1'>monomers</scene>,that create ring-shaped histone chaperone. The monomers are formed by a <scene name='46/467273/Core_domain/1'>core domain</scene> that responsible for oligomerization, that make the protein highly stable and compact. | ||
| + | The activation of NP is by strong destabilization of the pentamer, probably due to electrostatic repulsion. The NP needs compact and stable structure so he can accumulate negative charges that weakens its quaternary interactions, and its required for its biological function. | ||
==3D structures of nucleoplasmin== | ==3D structures of nucleoplasmin== | ||
| - | |||
[[1k5j]] – XlNpl core – ''Xenopus laevis'' | [[1k5j]] – XlNpl core – ''Xenopus laevis'' | ||
| Line 21: | Line 24: | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | 2. Taneva SG1, Muñoz IG, Franco G, Falces J, Arregi I, Muga A, Montoya G, Urbaneja MA, Bañuelos S. Activation of nucleoplasmin, an oligomeric histone chaperone, challenges its stability.Biochemistry. 2008 Dec 30;47(52):13897-906. PMID:19055325 DOI:10.1021/bi800975r | ||
| + | |||
| + | 3. Dutta S1, Akey IV, Dingwall C, Hartman KL, Laue T, Nolte RT, Head JF, Akey CW. The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly. Mol Cell.2001 Oct;8(4):841-53. PMID:11684019 | ||
| + | 4. SHU Te-Jun, ZHANG Yao-Zhou. Nucleoplasmin, an Important Nuclear Chaperone. Chinese Journal of Biochemistry and Molecular Biol. 15 March 2007 http://cjbmb.bjmu.edu.cn/EN/Y2007/V23/I09/718 | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Revision as of 11:02, 24 February 2018
| |||||||||||