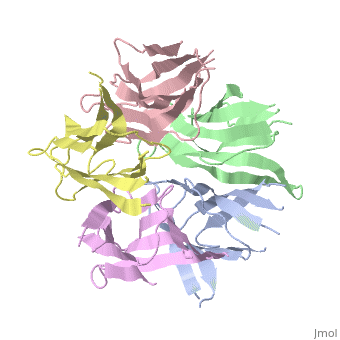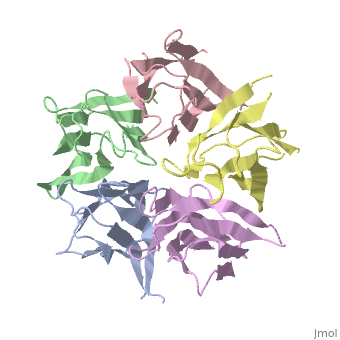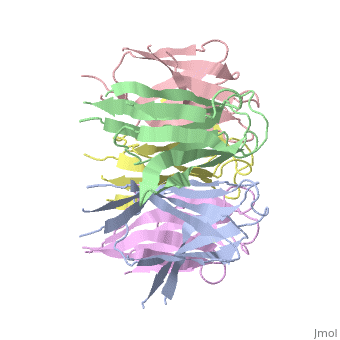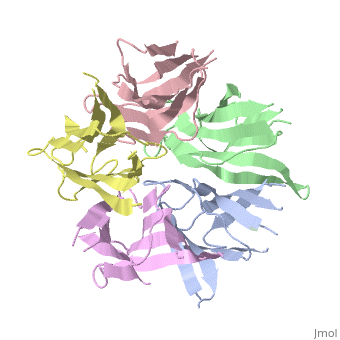This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Nucleoplasmin
From Proteopedia
(Difference between revisions)
| Line 12: | Line 12: | ||
The activation of NP is by strong destabilization of the pentamer, probably due to electrostatic repulsion. The NP needs compact and stable structure so he can accumulate negative charges that weakens its quaternary interactions, and its required for its biological function. | The activation of NP is by strong destabilization of the pentamer, probably due to electrostatic repulsion. The NP needs compact and stable structure so he can accumulate negative charges that weakens its quaternary interactions, and its required for its biological function. | ||
| + | = orthologey in Homo sapiens = | ||
| + | the NP in Humans is made out of <scene name='46/467273/Dimer/1'>dimer</scene> while each <scene name='46/467273/Monomer/1'>monomer</scene> consisted of five chains. The structure remain similiar to the one in Xenopus but with a change in amino acids in its core domain, Val insted of Ile. | ||
| + | The Decamer bind H2A-H2B dimers and H3-H4 tetramers simultaneously, In the absence of histone tetramers the pentamer binds H2A-H2B and formes central hub. When H3-H4 tetramers are recruited this results in a functional dimerization of the complex, and the decamer being formed. | ||
==3D structures of nucleoplasmin== | ==3D structures of nucleoplasmin== | ||
| Line 23: | Line 26: | ||
[[1ee5]], [[1ejy]] –XlNpl nuclear localization peptide + karyopherin-a | [[1ee5]], [[1ejy]] –XlNpl nuclear localization peptide + karyopherin-a | ||
| + | [[3t30]] - Human nucleoplasmin | ||
==== References ==== | ==== References ==== | ||
<references/> | <references/> | ||
Revision as of 09:23, 2 March 2018
| |||||||||||