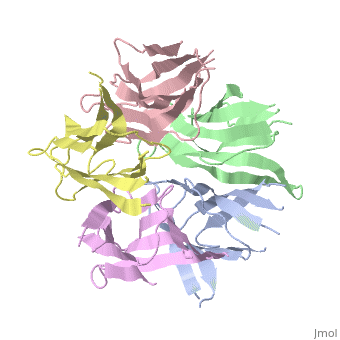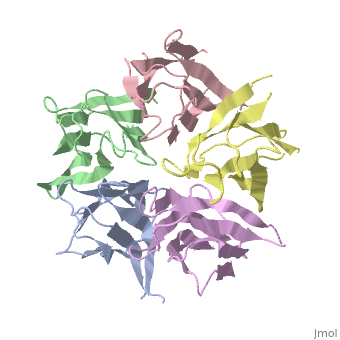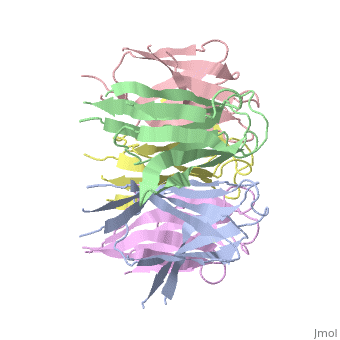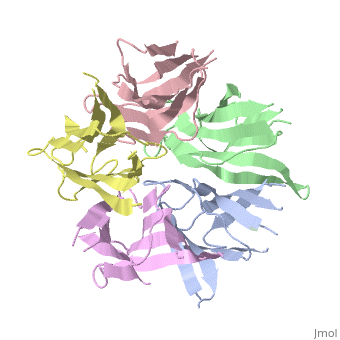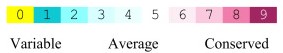We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Nucleoplasmin
From Proteopedia
(Difference between revisions)
| Line 10: | Line 10: | ||
=Structural highlights= | =Structural highlights= | ||
Nucleoplasmin (NP) is made out of five <scene name='46/467273/Np_monomer/1'>monomers</scene>,that create ring-shaped histone chaperone. The monomers are formed by a <scene name='46/467273/Core_domain/1'>core domain</scene> that responsible for oligomerization, that make the protein highly stable and compact. | Nucleoplasmin (NP) is made out of five <scene name='46/467273/Np_monomer/1'>monomers</scene>,that create ring-shaped histone chaperone. The monomers are formed by a <scene name='46/467273/Core_domain/1'>core domain</scene> that responsible for oligomerization, that make the protein highly stable and compact. | ||
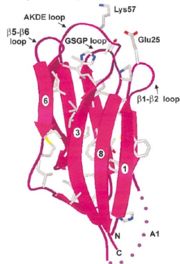
| - | The NP core is made out of eight | + | The NP core is made out of eight β strands that form a barrel with a jellyroll topology. Residues in the hydrophobic core are highly conserved (aproximate 80%). The AKDE and GSGP motifs are ordered loops and may function in decamer formation. |
[[Image:NP core.jpg|thumb|NP core]] | [[Image:NP core.jpg|thumb|NP core]] | ||
| Line 56: | Line 56: | ||
the NP in Humans is made out of <scene name='46/467273/Dimer/1'>dimer</scene> while each <scene name='46/467273/Monomer/2'>monomer</scene> consisted of five chains. The structure remain similiar to the one in Xenopus but with a change in amino acids in its <scene name='46/467273/Core_domain/2'>core domain</scene>, Val insted of Ile. | the NP in Humans is made out of <scene name='46/467273/Dimer/1'>dimer</scene> while each <scene name='46/467273/Monomer/2'>monomer</scene> consisted of five chains. The structure remain similiar to the one in Xenopus but with a change in amino acids in its <scene name='46/467273/Core_domain/2'>core domain</scene>, Val insted of Ile. | ||
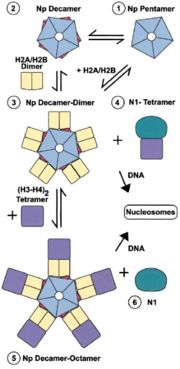
The Decamer bind H2A-H2B dimers and H3-H4 tetramers simultaneously, In the absence of histone tetramers the pentamer binds H2A-H2B and formes central hub. When H3-H4 tetramers are recruited this results in a functional dimerization of the complex, and the decamer being formed. | The Decamer bind H2A-H2B dimers and H3-H4 tetramers simultaneously, In the absence of histone tetramers the pentamer binds H2A-H2B and formes central hub. When H3-H4 tetramers are recruited this results in a functional dimerization of the complex, and the decamer being formed. | ||
| + | [[Image:Histone Storage and Nucleosome Assembly.jpg|thumb|Histone Storage and Nucleosome Assembly]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
== conservastion == | == conservastion == | ||
| Line 106: | Line 139: | ||
6. Sonia Ban˜ uelos,Miren J. Omaetxebarria, Isbaal Ramos,Martin R. Larsen, Igor Arregi, Ole N. Jensen, Jesus M. Arizmendi, Adelina Prado, and Arturo Muga. Phosphorylation of Both Nucleoplasmin Domains Is Required | 6. Sonia Ban˜ uelos,Miren J. Omaetxebarria, Isbaal Ramos,Martin R. Larsen, Igor Arregi, Ole N. Jensen, Jesus M. Arizmendi, Adelina Prado, and Arturo Muga. Phosphorylation of Both Nucleoplasmin Domains Is Required | ||
for Activation of Its Chromatin Decondensation Activity. THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 282, NO. 29, pp. 21213–21221, July 20, 2007 | for Activation of Its Chromatin Decondensation Activity. THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 282, NO. 29, pp. 21213–21221, July 20, 2007 | ||
| + | |||
| + | 7. Shuchismita Dutta, Ildiko´ V. Akey,Colin Dingwall, Kari L. Hartman,Tom Laue, Robert T. Nolte,James F. Head, and Christopher W. Akey. The Crystal Structure of Nucleoplasmin-Core: | ||
| + | Implications for Histone Binding and Nucleosome Assembly. Molecular Cell, Vol. 8, 841–853, October, 2001 | ||
Revision as of 16:51, 11 March 2018
| |||||||||||