Pho4
From Proteopedia
m |
|||
| Line 6: | Line 6: | ||
Phosphate system positive regulatory protein '''Pho4''' is a transcriptional activator which regulates the expression of phosphatase under conditions of phosphate starvation<ref>PMID:2220078</ref>. For details see [[Pho4 bHLH Protein]]. | Phosphate system positive regulatory protein '''Pho4''' is a transcriptional activator which regulates the expression of phosphatase under conditions of phosphate starvation<ref>PMID:2220078</ref>. For details see [[Pho4 bHLH Protein]]. | ||
*<scene name='72/725407/Cv/2'>Yeast Pho4 DNA-binding domain interactions with DNA</scene>. | *<scene name='72/725407/Cv/2'>Yeast Pho4 DNA-binding domain interactions with DNA</scene>. | ||
| + | Its structure is a basic basic helix-loop-helix (bHLH) [1] | ||
| + | The pho4 protein takes part in transcription of several yeast genes related to phosphate metabolism: | ||
| + | PH05- encoding repressible acid phosphatase [2] | ||
| + | PH08-encoding alkaline phosphatase [2] | ||
| + | PH081-a regulatory gene [3] | ||
| + | PH084- phosphate permease gene [4] | ||
| + | |||
</StructureSection> | </StructureSection> | ||
| + | ==sturcture== | ||
| + | |||
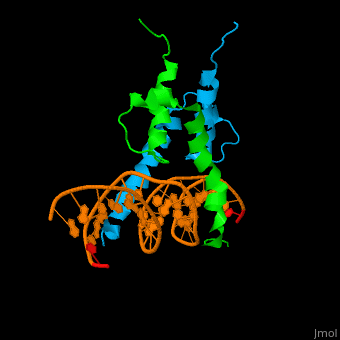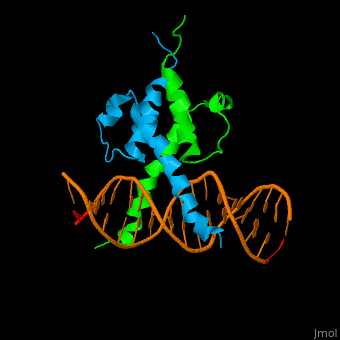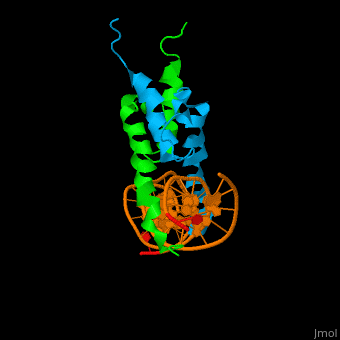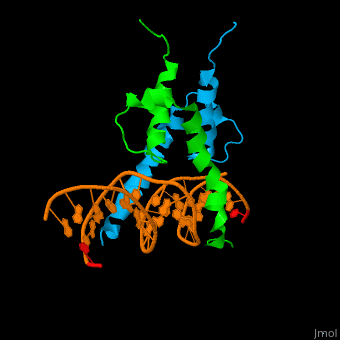
| + | The structure of the Pho4 protein is a basic Helix-loop-helix(bHLH) , The linker sequence between Helix1(H1) and Helix2(H2) contains several residues: Asn, Glu, Pro and Ser[6]. The DNA binding site is a homodimer and two monomers in a left‐handed four‐helix bundle fold (identical to bHLH/Zip and bHLH proteins fold) [7]. | ||
| + | |||
| + | [[loop structure]] | ||
| + | Pho4 loops (conserved structure?) are long but compact and forms a short alpha-helical structure [5]. The PHO4 loop contains a Trp residue that faces the aromatic rings of Tyr52 and His55 of helix H2, and Pro28 of H2 and H1.an aromatic cluster forms a cap structure is observed. | ||
| + | |||
== 3D Structures of lactoferrin == | == 3D Structures of lactoferrin == | ||
| Line 18: | Line 32: | ||
== References == | == References == | ||
<references/> | <references/> | ||
| - | [[Category:Topic | + | [[Category:Topic –Page]] |
Revision as of 12:13, 19 March 2018
| |||||||||||
sturcture
The structure of the Pho4 protein is a basic Helix-loop-helix(bHLH) , The linker sequence between Helix1(H1) and Helix2(H2) contains several residues: Asn, Glu, Pro and Ser[6]. The DNA binding site is a homodimer and two monomers in a left‐handed four‐helix bundle fold (identical to bHLH/Zip and bHLH proteins fold) [7].
loop structure Pho4 loops (conserved structure?) are long but compact and forms a short alpha-helical structure [5]. The PHO4 loop contains a Trp residue that faces the aromatic rings of Tyr52 and His55 of helix H2, and Pro28 of H2 and H1.an aromatic cluster forms a cap structure is observed.
3D Structures of lactoferrin
Updated on 19-March-2018
1a0a – yPho4 DNA-binding domain + DNA - yeast
3w3x – yPho4 peptide + importin subunit b-3
References
- ↑ Berben G, Legrain M, Gilliquet V, Hilger F. The yeast regulatory gene PHO4 encodes a helix-loop-helix motif. Yeast. 1990 Sep-Oct;6(5):451-4. PMID:2220078 doi:http://dx.doi.org/10.1002/yea.320060510
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Inbar Yaffe, Alexander Berchansky, Joel L. Sussman